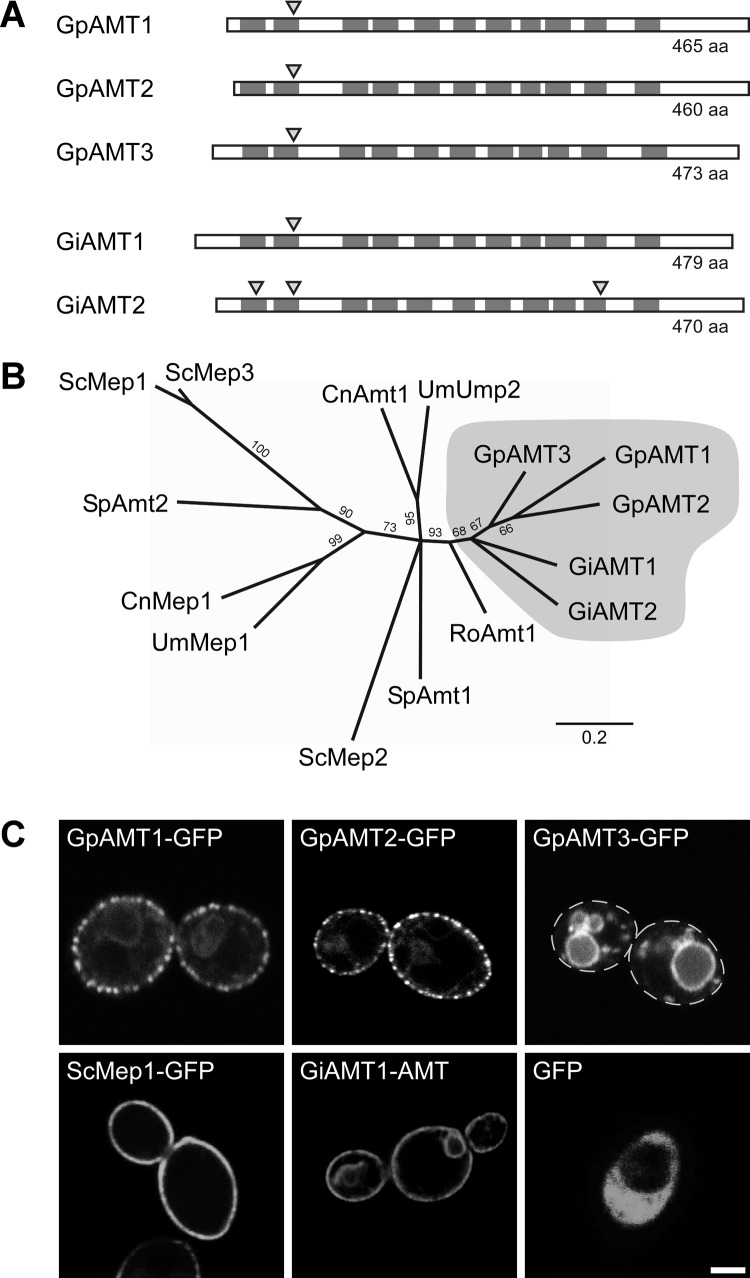
Fig 1.
Characteristics of Geosiphon pyriformis ammonium transporters (AMTs). (A) Transmembrane domain (TMD) topology and intron localization in comparison with those of the Rhizophagus irregularis ammonium transporters GiAMT1 (CAI54276) and GiAMT2 (CAX32490). Dark gray boxes indicate the positions of TMDs, and triangles mark intron positions. TMD positions and the position of one intron are highly conserved, while N and C termini differ in length and are less conserved. Schematic is to scale. aa, amino acids. (B) Unrooted phylogenetic tree computed by RAxML maximum-likelihood analysis (1,000 bootstraps) of selected representative fungal ammonium transporters. Branches with bootstrap support below 60% were collapsed to polytomies. Sequences with the following accession numbers were obtained from the GenBank database: Schizosaccharomyces pombe SpAmt1, NP_588424, and SpAmt2, XP_002175812; Saccharomyces cerevisiae ScMep1, P40260, ScMep2, P41948, and ScMep3, P53390; Ustilago maydis UmMep1, AAL08424, and UmUmp2, AAO42611; Cryptococcus neoformans CnMep1, XP_566614, and CnAmt1, XP_567361; and Rhizopus oryzae RoAmt1, EIE87911. Bootstrap support is indicated at the branches, scale bar indicates substitutions per site, and gray shading marks glomeromycotan AMTs. (C) Heterologous expression of soluble GFP or GFP-tagged ammonium transporter proteins in the ammonium transporter-deficient S. cerevisiae mutant strain MLY131a/α. Confocal laser-scanning microscopy (CLSM) results are shown; dashed lines in GpAMT3-GFP image indicate cell borders. Bar, 2 μm.