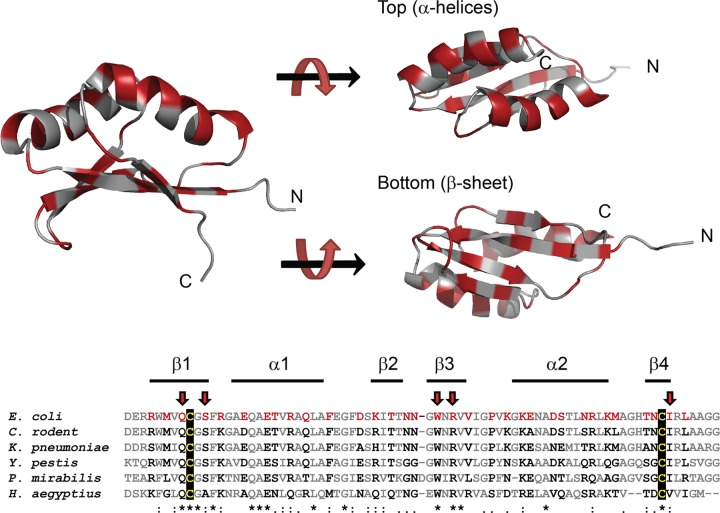
Fig 2.
Locations of FtsNSPOR amino acids targeted for mutagenesis. (Top) Ribbon diagrams illustrating the locations of all residues (red) where amino acid substitutions were introduced to test for localization defects. (Bottom) Alignment of the SPOR domains from several FtsN proteins. Residues targeted for mutagenesis are shown in red, with arrows pointing to residues where substitutions impaired localization at least 3-fold. The black background highlights the two conserved cysteines proposed to form a disulfide bond. The alignment was prepared in Clustal W (38) with the following FtsN SPOR domain sequences: Escherichia coli MG1655 (locus tag b3833) residues 244 to 319, Citrobacter rodentium ICC168 (ROD_38131) residues 236 to 310, Klebsiella pneumoniae 342 (KPK_5451) residues 245 to 320, Yersinia pestis bv. Antiqua B42003004 (YpAngola_A0114) residues 206 to 281, Proteus mirabilis ATCC 29906 (HMPREF0693_0033) residues 188 to 264, and Haemophilus aegyptius ATCC 11116 (HMPREF9095_0699) residues 185 to 256.