Fig 4.
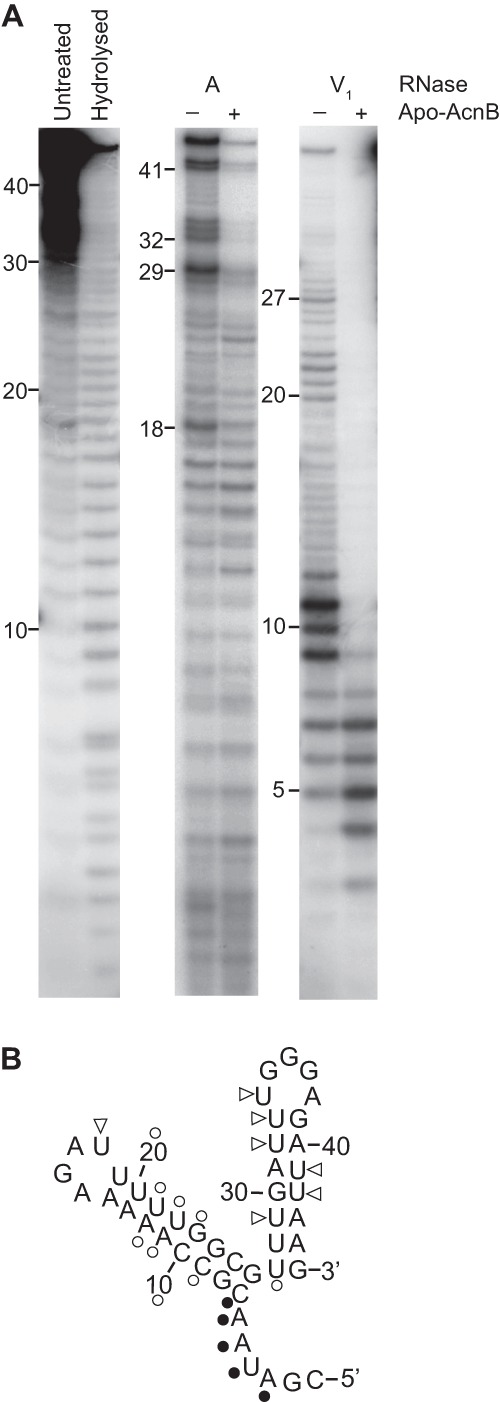
RNA footprinting. (A) 5′-End-labeled 45-nt pgdA RNA was incubated with (+) and without (−) apo-AcnB and subjected to RNase digestion. (B) RNase A (triangles) and RNase V1 (circles) cleavage sites were mapped on the predicted pgdA 3′ UTR structure. Open symbols indicate nucleotides that were more protected from RNase cleavage in the presence of apo-AcnB, whereas filled symbols indicate those nucleotides that were less protected.
