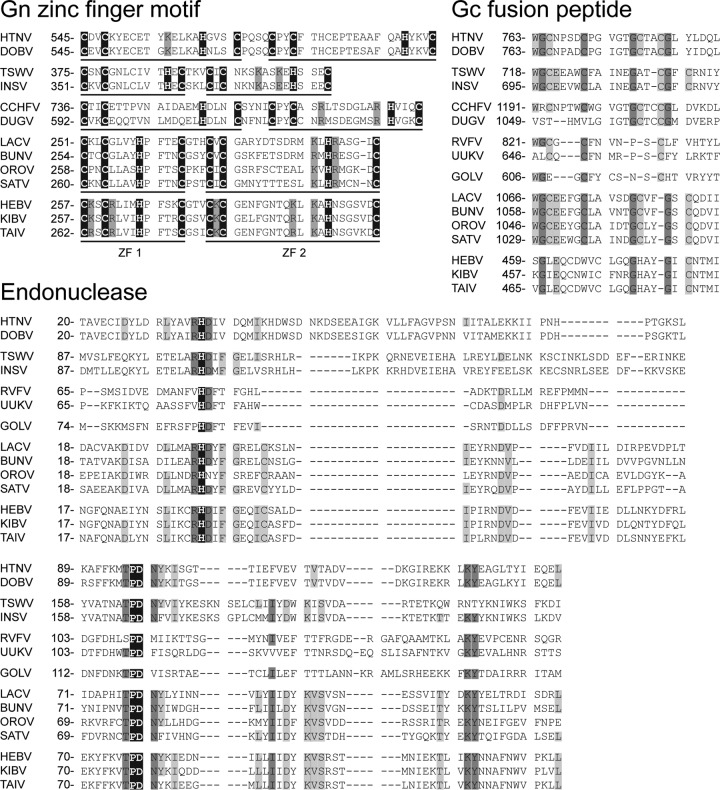
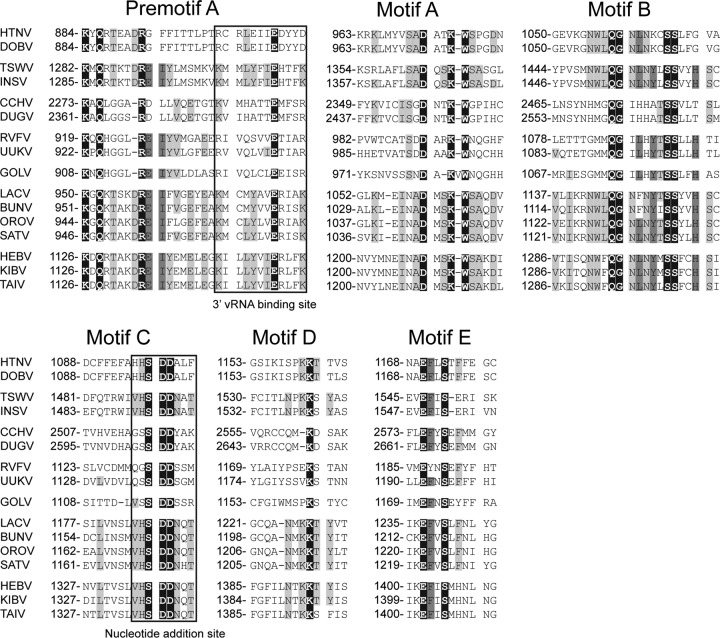
Fig 5.
Multiple-sequence alignments of conserved domains of HEBV, TAIV, KIBV, and other bunyaviruses. Alignments were performed by using the E-INS-I algorithm in MAFFT and manually edited. Numbers represent genome positions. Amino acids with 100% identity are highlighted in black, those with 75% identity are highlighted in dark gray, and those with 50% identity are highlighted in light gray. Gn zinc finger motifs are highlighted in black, and conserved basic residues are highlighted in dark gray.