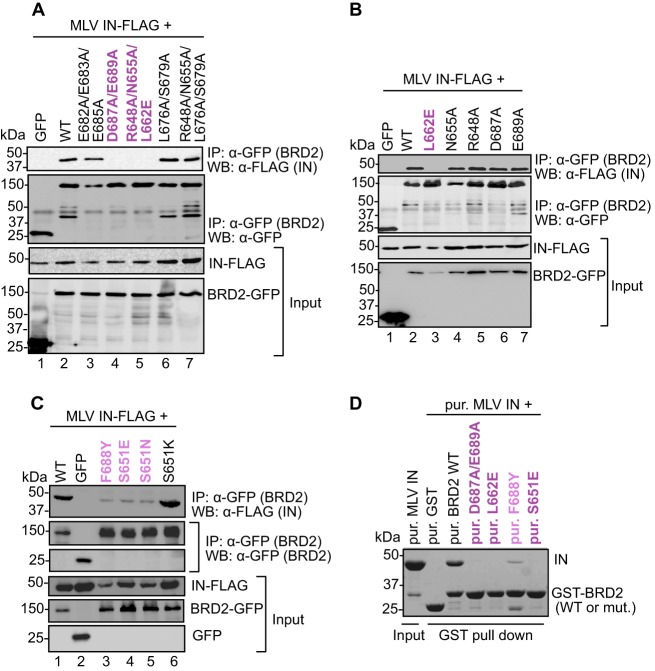
Fig 2.
Mapping the IN binding interface in the BRD2 ET domain. (A to C) Coimmunoprecipitation of FLAG-tagged MLV IN with GFP-tagged WT BRD2 or its point mutants. Proteins recovered after coimmunoprecipitation with a GFP affinity matrix were analyzed by Western blotting using mouse anti-GFP or anti-FLAG antibodies as indicated. (D) GST pulldown assay. GST-tagged BRD2(641–710) or its point mutants were incubated with MLV IN and glutathione Sepharose beads. Proteins bound to beads were separated by 12% denaturing PAGE and detected by staining with Coomassie blue. Residues whose mutations abrogate binding are colored dark purple; residues whose mutations displayed weaker binding to MLV IN are shown in light purple.