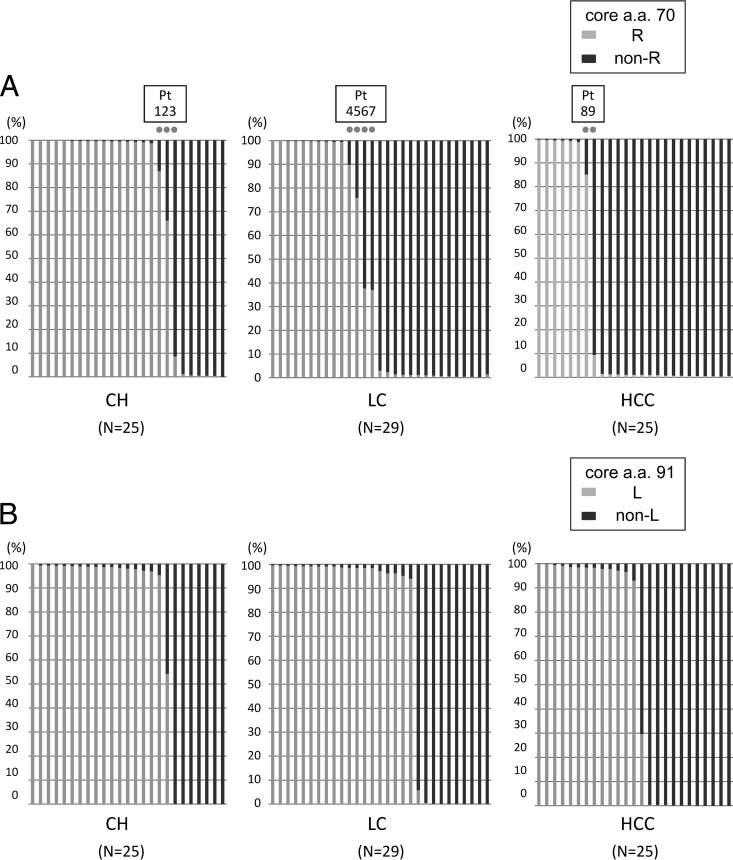
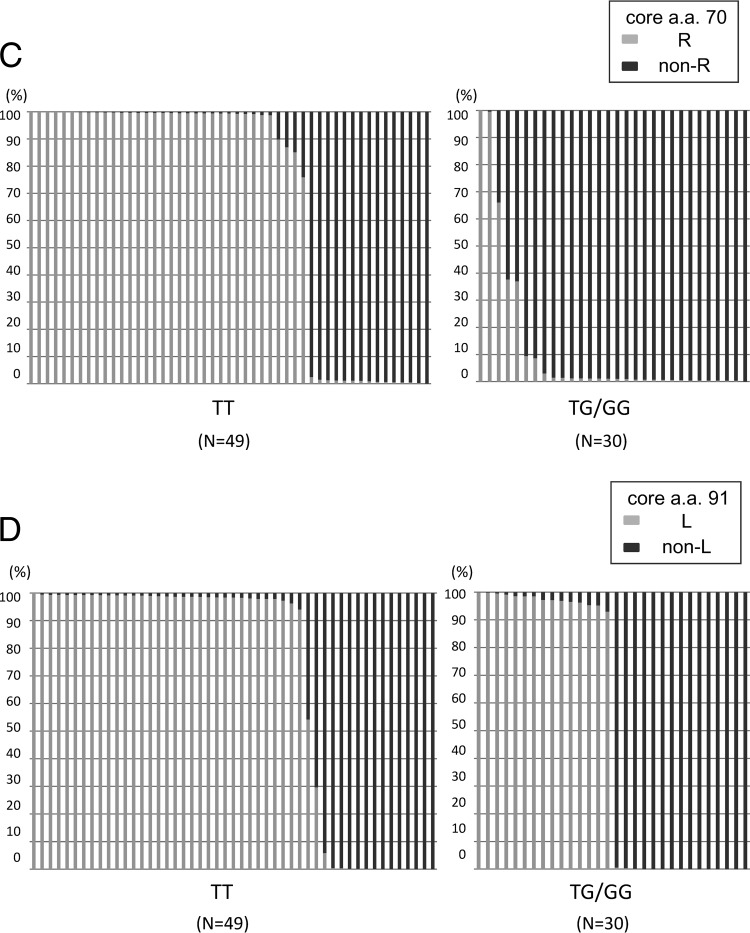
Fig 2.
The core regions of the HCV genomes from 79 patients persistently infected with HCV genotype 1b (25 patients with chronic hepatitis [CH], 29 with liver cirrhosis [LC], and 25 with hepatocellular carcinoma [HCC]) were subjected to deep sequencing. Each bar represents the result for a single patient. At the specific locations of core aa 70 and aa 91, no single nucleotide mutation was observed in the previous control plasmid experiment. (A) Disease stages and percentages of mutations at core aa 70. Nine dots indicate the nine patients with a high mixture rate (between 5% and 95%) at core aa 70 (R and non-R). (B) Disease stages and percentages of mutations at core aa 91. (C) IL28B SNP and percentages of mutations at core aa 70. (D) IL28B SNP and percentages of mutations at core aa 91.