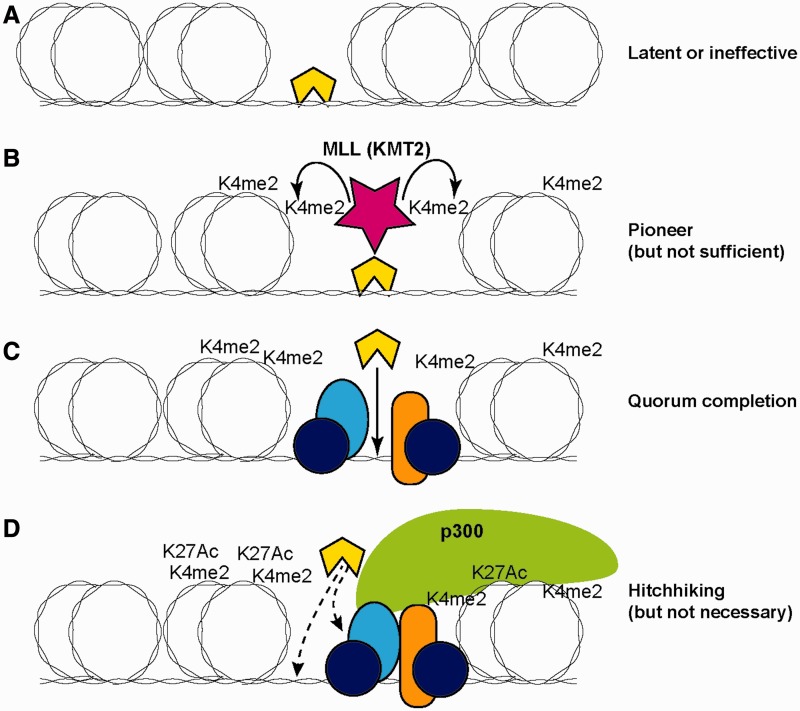
Figure 1:
Functional modes of transcription factor engagement in a chromatin context. The transcription factor of interest is represented by a yellow (light color) polygon. Double coils around an oval represent DNA wrapped around nucleosomes. (A) Binding alone to a temporarily accessible site, without activation of histone modifications or redistribution. (B) Binding stably enough to recruit local histone modifying enzymes (MLL = KMT2, histone H3 Lys 4 methyltransferase) and enhance access to DNA, thus providing a local pioneering effect even if not conferring immediate transcriptional regulatory function. K4me2 = H3K4me2. (C) Binding to complete a ‘regulatory quorum’. Factor of interest is selectively recruited to join a pre-established assemblage of other transcription factors at a cis-regulatory site that was poised but not functionally active until the factor joined. Recruitment in this case is shown by protein–DNA binding, but may also include protein–protein binding. (D) ‘Hitchhiking’: factor binding adventitiously to a cis-regulatory element at which function is already established independently of the factor of interest, and unaffected by the presence of the factor of interest. Factor may bind to exposed DNA sites or through interaction with other recruited proteins or histone marks. Element diagrammed is an activating enhancer element where H3K27Ac (H3K27Ac) modifications have occurred and the p300 co-activator has been recruited already; however, hitchhiking can also occur at active promoters where the marks are different.