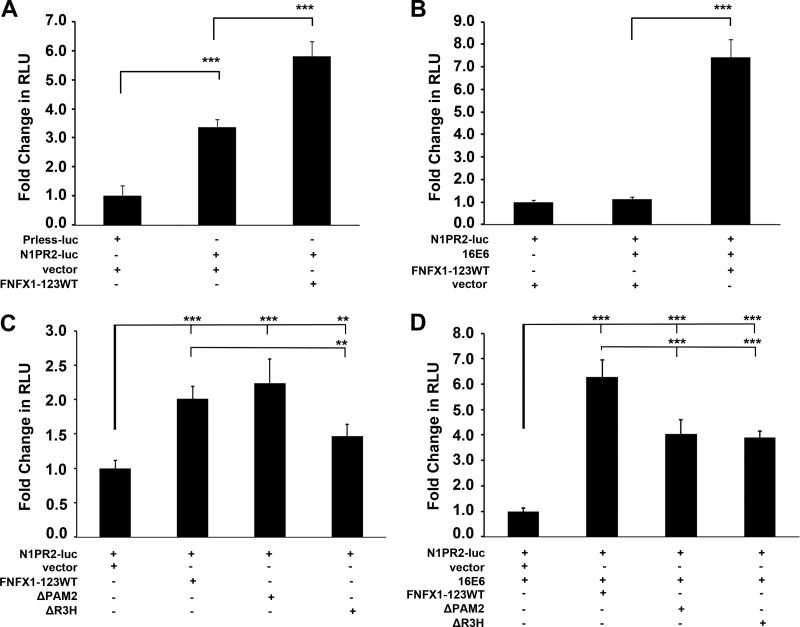
Fig 5.
Notch1 promoter-driven expression increased by coexpression of FNFX1-123WT and 16E6. (A) HFKs were transfected with either a promoterless luciferase construct (Prless-luc) or a 2-kb Notch1 promoter luciferase construct (N1PR2) and cotransfected with the FLAG-tagged NFX1-123 wild-type construct (FNFX1-123WT) or the pDEST12.2 vector (vector). N1PR2 produced significantly more baseline luciferase activity (RLU, relative light units) than Prless-luc. ***, P < 0.001. HFKs cotransfected with N1PR2 and FNFX1-123WT expressed more luciferase than N1PR2 and vector. ***, P < 0.001. (B) HFKs were cotransfected with N1PR2 and either vector, 16E6, or 16E6 and FNFX1-123WT. Eightfold more luciferase was produced when HFKs were cotransfected with N1PR2, 16E6, and FNFX1-123WT than with N1PR2, 16E6, and vector. ***, P < 0.001. (C) HFKs were cotransfected with N1PR2 and either the FNFX1-123WT construct, the FLAG-tagged NFX1-123 PAM2-deleted construct (ΔPAM2), the FLAG-tagged NFX1-123 R3H-deleted construct (ΔR3H), or vector. FNFX1-123WT had a moderate increase in luciferase compared to vector alone. ***, P < 0.001. A reduction in luciferase was detected when the R3H domain was removed from FNFX1-123WT. **, P < 0.01. (D) HFKs were cotransfected with N1PR2 and 16E6 and either FNFX1-123WT, ΔPAM2, ΔR3H, or vector. Sixfold more luciferase was produced when HFKs were cotransfected with N1PR2, 16E6, and FNFX1-123WT than with N1PR2, 16E6, and vector. ***, P < 0.001. Luciferase was significantly decreased when the PAM2 motif or R3H domain was removed from FNFX1-123WT in 16E6-cotransfected HFKs. ***, P < 0.001. All graphs are of at least three combined experiments, each conducted in triplicate. Error bars graphed represent 95% confidence intervals, and P values were calculated by two-tailed student t test with vector as the reference.