Fig 4.
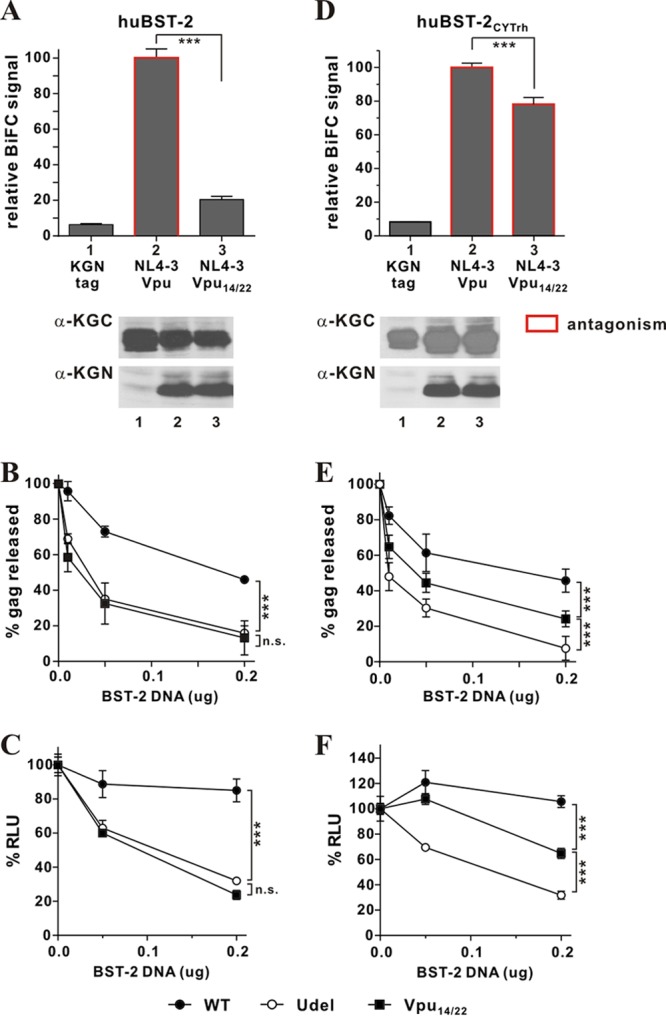
Gain-of-function analysis of cytoplasmic interactions between Vpu and BST-2. (A to C) Mutation of residues 14 and 22 in the TM domain of NL4-3 Vpu results in functional inactivation. (A) The physical interaction of huBST-2 and Vpu14/22 was quantified by flow cytometry as described for Fig. 1B. The BiFC signal measured for the interaction of wild-type Vpu and huBST-2 (bar 2) was defined as 100%. ***, P < 0.01. The expression of KGC-tagged huBST-2 and KGN-tagged proteins was assessed by immunoblotting as described for Fig. 1B. The KGN fragment encoded by the “tag-only” vector has a faster mobility and is not visible here. Faint bands in lane 1 are nonspecific background. (B) The functional antagonism of human BST-2 by Vpu variants was assessed by metabolic labeling as described for Fig. 3C. Filled circles, NL4-3 (WT); filled squares, NL4-3 Vpu14/22; open circles, NL4-3/Udel. Data are means plus standard errors of the means from three independent experiments. Statistical significance was determined by two-way ANOVA. ***, P < 0.01; n.s., P > 0.05. (C) The functional antagonism of human BST-2 by Vpu variants was assessed by a TZM-bl assay. 293T cells (5 × 106) were transfected with 5 μg of either pNL4-3 (WT), pNL4-3/Udel, or pNL4-3 Vpu14/22 in the absence of BST-2 or in the presence of 0.05 μg or 0.2 μg of pcDNA-huBST-2. The TZM-bl assay was carried out as described in Materials and Methods. Virus release in the absence of BST-2 (0 μg) was defined as 100%. Data are means plus standard errors of the means from three independent experiments. Statistical significance was determined by two-way ANOVA. ***, P < 0.01; n.s., P > 0.05. (D to F) Cytoplasmic interactions are sufficient to restore the BST-2-antagonizing activity of Vpu14/22. (D) The physical interaction between huBST-2CYTrh and Vpu or Vpu14/22 was quantified as described for Fig. 1B. The BiFC signal obtained for the interaction of NL4-3 Vpu and huBST-2CYTrh (bar 2) was defined as 100%. ***, P < 0.01. Bars outlined in red represent combinations where the respective Vpu variant was able to antagonize huBST-2CYTrh (as assessed in panels E and F). The expression of KGC-tagged huBST-2CYTrh and KGN-tagged proteins was assessed by immunoblotting as described for Fig. 1B. (E) The ability of Vpu14/22 to antagonize huBST-2CYTrh via cytoplasmic domain interaction was assessed by metabolic labeling as described for Fig. 3C. Data are means plus standard errors of the means from three independent experiments. Statistical significance was determined by two-way ANOVA. ***, P < 0.01. (F) The ability of Vpu14/22 to antagonize huBST-2CYTrh was also assessed by a TZM-bl assay as described for panel C. Data are means plus standard errors of the means from three independent experiments. Statistical significance was determined by two-way ANOVA. ***, P < 0.01.
