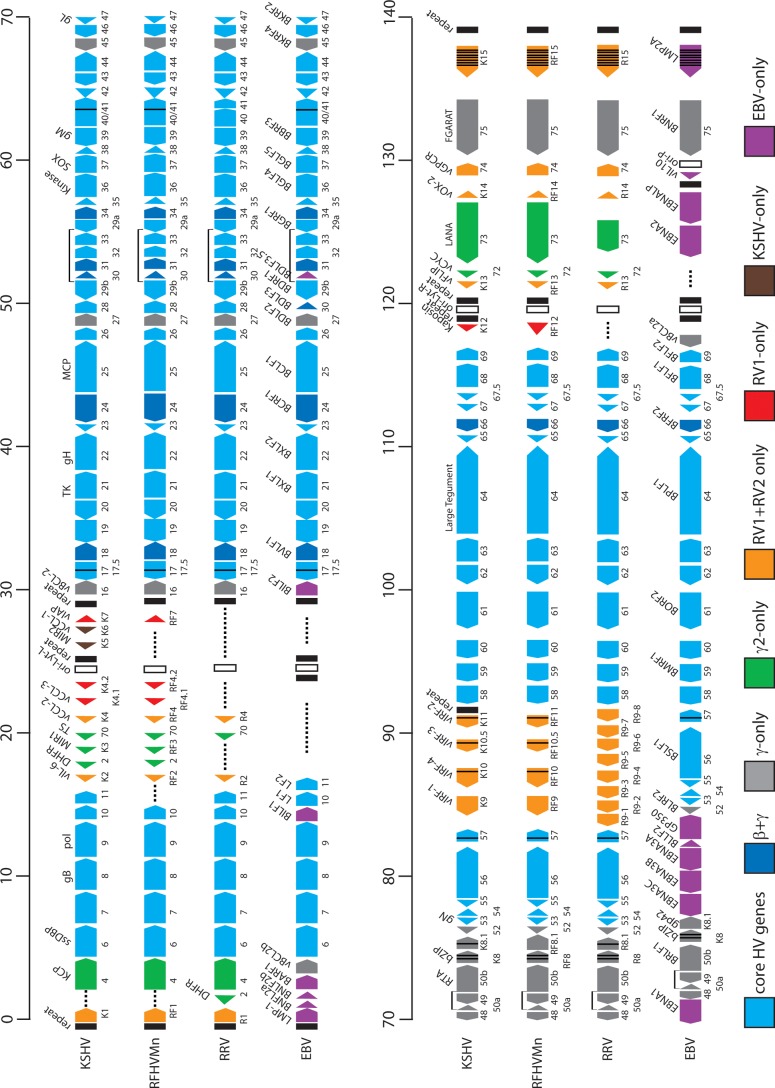
Fig 1.
Comparative map of the rhadinovirus and lymphocryptovirus genomes. The positions and transcription directions of the ORFs identified in the RFHVMn genome (KF703446; this study) are compared to the corresponding ORFs in KSHV (NC_009333), RRV (AF210726 and NC_003401), and EBV (NC_007605), as identified in the NCBI accession records (the basic design of this map was patterned after Fig. 1 of reference 22). The numbering of the KSHV, RFHVMn, and RRV ORFs is patterned after the genome structure of the prototype rhadinovirus, HVS, with KSHV–specific ORFs designated K1 to K15 and their homologs in RFHVMn and RRV designated RF1 to RF15 and R1 to R15, respectively. The EBV ORFs are indicated, using the typical EBV nomenclature above the ORF and the corresponding rhadinovirus designation below. The approximate positions of the ORFs are shown with regard to their positions within the KSHV genome (i.e., 1 to 140 kb). The sizes of the ORF markers are approximately consistent with the sizes of the actual encoded proteins. Vertical black lines within an ORF indicate splicing events or internal initiations, while longer–range splices of protein–coding exons are indicated with bars. The ORFs are color coded with regard to their conservation in other herpesvirus genomes, showing core HV genes conserved in most herpesvirus families, genes conserved in beta– and gammaherpesviruses, gammherpesviruses only, gamma–2–herpesviruses, RV1 and RV2 rhadinoviruses, RV1 rhadinoviruses only, KSHV only, and EBV only, as indicated. The RRV nomenclature from AF210726 was used for consistency.