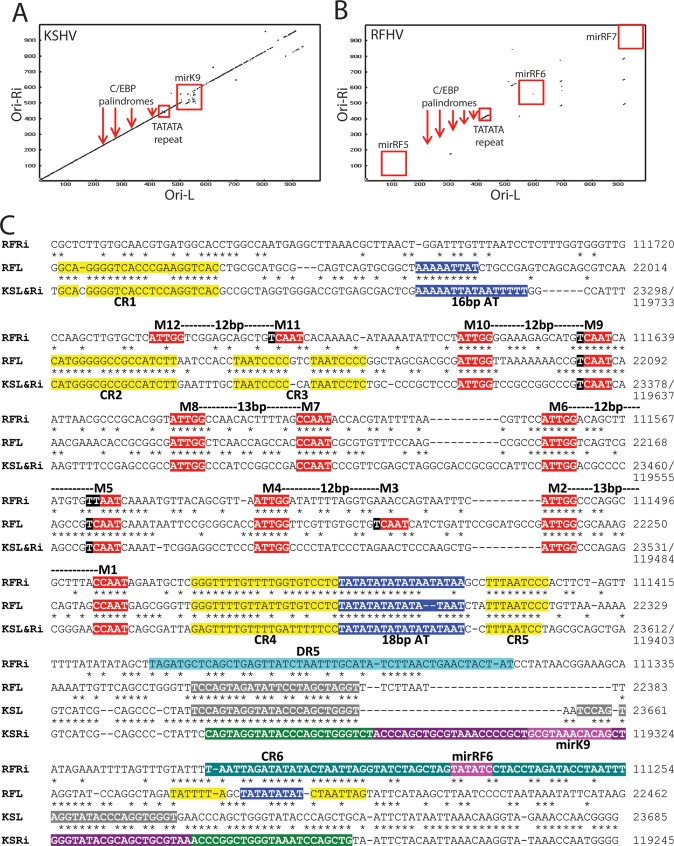
Fig 8.
Comparison of the long-inverted-repeat regions of Ori-Lyt-R and Ori-Lyt-L of KSHV and RFHVMn. (A and B) Dot plot of the nucleotide sequence alignment of the inverted-repeat regions of Ori-Lyt-L and Ori-Lyt-R for KSHV (A) and RFHVMn (B), with the approximate positions of the C/EBP palindromes (M1/2, M5/6, M7/8, and M9/10), the 18-bp TATATA repeat, and adjacent microRNA genes, illustrated in panel C. (C) Alignment of the nucleotide sequences of KSHV (U75698) containing the Ori-Lyt-L (bp 23223 to 23685) (KSL), the inverted sequence of KSHV containing the Ori-Lyt-R region bp (119245 to 119782) (KSRi), the nucleotide sequences of RFHVMn (KF703446; this study) containing the putative Ori-Lyt-L (bp 21937 to 22462) (RFL), and the inverted sequence of RFHVMn containing the putative Ori-Lyt-R (bp 111254 to 111800) (RFRi). The conserved AT-rich motifs (blue) and C/EBP motifs (red) are indicated (see the text). Other conserved regions (CR1 to CR6) are highlighted in yellow or gray. The kshv-miR-K12-K9 microRNA with the mature microRNA (purple) and stem-loops (green) and the predicted rfhvmn-miRc-RF6 are shown.