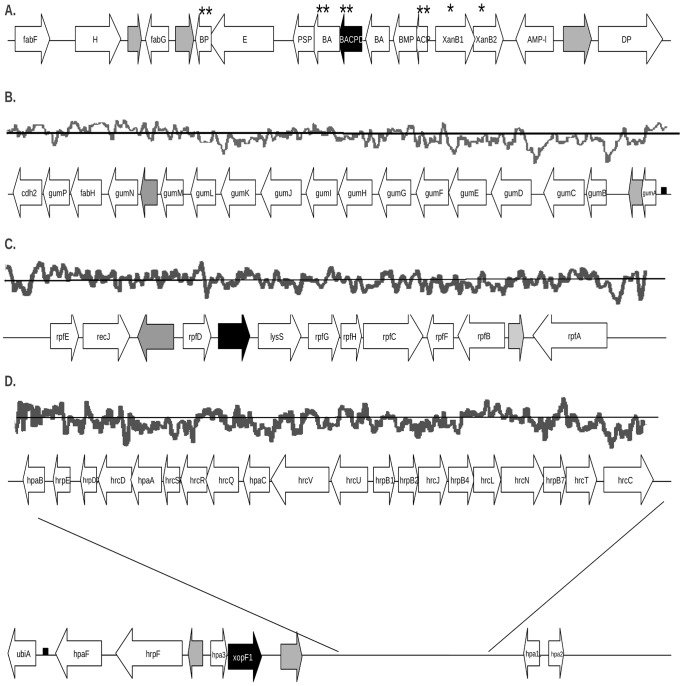
Figure 3. Organization of pathogenicity-related gene clusters in the Xam CIO151 genome.
Open arrows with labels indicate genes with assigned functions, black arrows indicate genes with early stop codons, open arrows without labels indicate conserved hypothetical proteins and grey arrows indicate non-conserved hypothetical proteins. Graphs above clusters show the G+C content and deviations from the average value. A. Xanthomonadin gene cluster; * indicates genes related to pigB genomic region and ** indicates genes reported as important for cluster functionality. Abbreviations used are: H = halogenase (xanmn_chr15_0075), BP = xanthomonadin biosynthesis protein (xanmn_chr15_0079), E = xanthomonadin exporter (xanmn_chr15_0373), PSP = putative secreted protein (xanmn_chr15_0080), BACPD = xanthomonadin biosynthesis acyl carrier protein dehydratase (xanmn_chr15_0082), BA = putative xanthomonadin biosynthesis acyltransferase (xanmn_chr15_0081 and xanmn_chr15_0083), BMP = putative xanthomonadin biosynthesis membrane protein (xanmn_chr15_0084), ACP = acyl carrier protein (xanmn_chr15_0085), XanB1 = putative reductase/halogenase (xanmn_chr15_0086), XanB2 = putative pteridine-dependent deoxygenase like protein (xanmn_chr15_0087), AMP-l = AMP-ligase (xanmn_chr15_0088), DP = dipeptidyl peptidase (xanmn_chr15_0090). B. Cluster implicated in xanthan production (gum). C. Regulation of pathogenicity factors (rpf) cluster. D. Type III secretion system (T3SS) cluster.