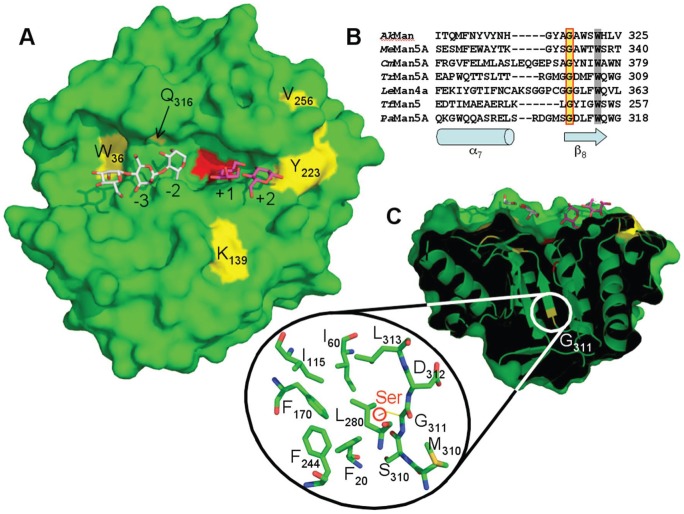
Figure 3. Structural view of PaMan5A (PDB 3ZIZ) exhibiting substituted amino-acids.
A. Surface view of the catalytic cleft of PaMan5A with mannotriose modelled in the −2 and −3 subsites and mannobiose modelled in the +1 and +2 subsites. The structures of GH5 from T. reesei and T. fusca in complex with mannobiose and mannotriose, respectively, were superimposed on the top of the structure of PaMan5A to map the substrate-binding subsites. The two catalytic glutamate residues, E177 and E283, are coloured in red. The substituted amino-acids are labelled and coloured in yellow. B. Structural based sequence alignment of the region around position 311 (according to PaMan5A numbering) from Podospora anserina (PaMan5A), Aplysia kurodai (AkMan, PDB 3VUP), Mytilus edulis (MeMan5A, PDB 2C0H), Cellvibrio mixtus (CmMan5A, PDB 1UUQ), Trichoderma reesei (TrMan5A, PDB 1QNR), Lycopersicon esculentum (LeMan4A, PDB 1RH9) and Thermomonospora fusca (TfMan5, PDB 2MAN). Secondary structure elements, α-helix α7 and β-strand β8, are indicated below the sequences as a cylinder and an arrow, respectively. Strictly conserved residues, G311 and W315 (according to PaMan5A numbering), are shown with a yellow and a grey background, respectively. C. Surface view of PaMan5A rotated of about 90° along the horizontal axis. The front clipping plane has been moved in order to visualize the location of G311 inside the molecule. The zoom shows a compact hydrophobic core in the vicinity of G311.