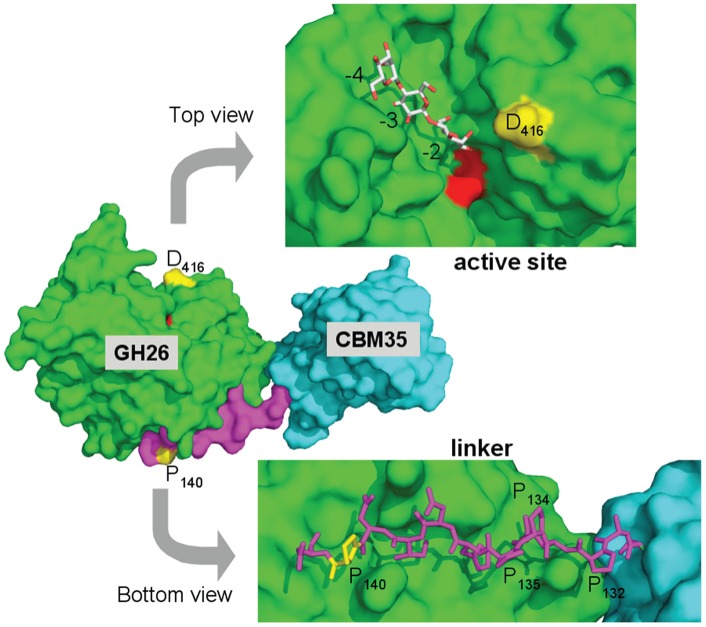
Figure 5. Structural view of PaMan26A (PDB 3ZM8) exhibiting substituted amino-acids.
The central panel shows a surface view of the entire PaMan26A structure, which is composed of a carbohydrate binding module (CBM) belonging to the CBM35 family in cyan, a linker in violet and a catalytic domain belonging to the GH26 family in green. The two catalytic glutamate residues, E300 and E390, are coloured in red. The two substituted amino-acids, P140 and D416, are labelled and coloured in yellow. The top view represents the surface view of the catalytic cleft of PaMan26A rotated about 90° along the horizontal axis with mannotriose modelled into the −2 to −4 subsites. The structure of GH26 from C. fimi in complex with mannotriose was superimposed on the top of the structure of PaMan26A to map the substrate-binding subsites. The bottom view displays the PaMan26A linker (from residue 131 to residue 141) in stick representation. The molecule has been rotated of about 90° along the horizontal axis and in the opposite direction compared to the top view. The proline residues of the linker are labelled.