Abstract
The objective of this study was to characterize the morphology, size-distribution, concentration and genome size of virus-like particles (VLPs) in two acetate-fed Methanosaeta-dominated reactors to better understand the possible correlation between viruses and archaeal hosts. The study reactors were dominated by a single genus of acetoclastic methanogen, Methanosaeta, which was present at 6 to 13 times higher than the combined bacterial populations consisting of Proteobacteria, Firmicutes, and Bacteroidetes. Epifluorescent microscopy showed VLPs concentration of 7.1 ± 1.5×107 VLPs/ml and 8.4 ± 4.3×107 VLPs/ml in the two laboratory reactors. Observations of no detectable import of VLPs with the reactor feed combined long operational time since the last inocula were introduced suggests that the VLP populations were actively propagating in the reactors. Transmission electron microscopy images showed VLPs with morphology consistent with Siphoviridae in both reactors, and VLPs with morphologies consistent with Myoviridae in one of the reactors. The morphology, size-distribution and genome size of VLPs were distinct between reactors suggesting that unique viral populations inhabited each reactor, though the hosts of these VLPs remain unclear.
Introduction
Acetoclastic methanogens inhabit a variety of natural ecosystems such as paddy field soils [1], fresh water and marine sediments [2,3] and acidic fens [4]. They are among the most important methane producers on earth, since two-thirds of the biogenic methane released to atmosphere is thought to be derived from the methyl group of acetate [5,6].
Most methanogens use hydrogen and carbon dioxide as substrates (hydrogenotrophic methanogens) while only two known genera of methanogens, Methanosaeta and Methanosarcina, are able to utilize acetate (acetoclastic methanogens). Because they use the same substrate, acetate, Methanosarcina and Methanosaeta compete with each other using different growth strategies. Having a higher growth rate (k) and lower acetate affinity (higher Ks), Methanosarcina usually dominate in environments with high acetate concentrations. In contrast, with lower growth rate and higher acetate affinity, Methanosaeta is generally dominant when acetate concentrations decrease. Methanosaeta is ubiquitous and dominant in major biogenic methane releasing environments and have been postulated to be the predominant methane producer on earth [5].
Acetoclastic methanogens are also essential in engineered systems such as anaerobic digestion. Anaerobic digestion is used worldwide to treat organic waste generated from municipal, industrial and agricultural sources. Stability of anaerobic digestion has been investigated and improved over several decades [7–10]. Nevertheless, upsets and failures of the process still occur with no obvious explanation. Digestion failure is usually characterized by a decreased rate of methane production, accumulation of acetic and short chain length organic acids, and decreased pH [11], which implies the loss of functional microorganisms involved in the last step of anaerobic digestion, which is methanogenesis.
Viruses of Bacteria (bacteriophages or phages) and Archaea (archeoviruses) [12], have been shown to influence the composition of microbial communities through cell lysis [13]. By targeting the most rapid growing populations (“kill the winner” theory), viruses are believed to stimulate microbial diversity [14]. Diverse and abundant virus-like particles (VLPs) have been reported to inhabit anaerobic digestion systems, suggesting that VLPs may play an important role in functioning of this treatment process [15,16]. About two-thirds of the methane produced in mesophilic anaerobic digesters is from acetoclastic methanogens [17,18], and Methanosaeta is typically the dominant genus [19], which suggesting Methanosaeta may be a favorable target for viral attack. Therefore, viral attack on the Methanosaeta may explain observed process upsets characterized by loss of methane production. Viruses of acetoclastic methanogens have not been isolated, a task which is complicated by slow host growth rates (4.8 day doubling time for Methanosaeta) [20] and lack of anaerobic solid media methods for plaque assays (i.e. one month of Methanosaeta growth results in <1 mm diameter colonies and no lawn formation [21]).
The goals of this study were (1) to investigate the occurrence and persistence of viruses in two Methanosaeta-dominated reactors (2), to examine the morphology, size distribution and genome length of observed viruses, and (3) to determine the concentration of viruses in relation to acetoclastic methanogens and bacteria using two previously-established acetate-fed methanogenic enrichment reactors.
Materials and Methods
Reactors Operation and Analyses
The study reactors consisted of two semi-continuous stirred tank reactors (CSTR) that were originally established in 2002, as described by Conklin et al [22]. The reactors were inoculated with anaerobic digester sludge from the West Point Treatment Plant in Seattle, WA, fed aseptic Reduced Anaerobic Mineral Media (RAMM) supplemented with vitamins and acetate (234mM) as the sole carbon and energy source, and operated at 30-34°C. One reactor was fed in a single daily dose (daily-fed reactor) and the other in hourly increments (hourly-fed reactor). The daily-fed reactor was re-inoculated in 2007 and 2008 due to community population shift from Methanosarcina to Methanosaeta. But reestablishment of Methanosarcina population did not succeed. Table 1 summarizes the operating conditions for the reactors during the current study. During the study period, the reactors were fed 150 ml/day sterile feed and the same volume was manually wasted daily from each reactor. Due to a lower operating reactor volume in the daily-fed reactor during the study period, this reactor had a lower hydraulic retention time (HRT) (13.3 days) than the hourly-fed reactor (20 days).
Table 1. Experimental reactor characteristics.
| Parameter | daily-fed reactor | hourly-fed reactor |
|---|---|---|
| reactor volume | 2 L | 3 L |
| vol. of media fed per day | 150 ml | 150 ml |
| hydraulic retention time | 13.3 days | 20 days |
| date of inoculation | March 2008 | January 2002 |
| pH | 7.3 | 7.4 |
| CH4 % in headspace | 54% | 56% |
Gas production by the reactors was continually recorded by wet test meters (Precision Scientific, Chicago, IL). The concentrations of methane and carbon dioxide in headspace gas were analyzed by Carle Series 100A gas chromatograph equipped with a thermal conductivity detector (GC-TCD, Chandler Engineering, Tulsa, OK). The concentration of acetate in the reactor effluent was analyzed using a Shimadzu GC-2010 equipped with flame ionization detector (GC-FID, Shimadzu Scientific Instruments, Columbia, MD) and a DB-FFAP column (122-3232, Agilent Technologies, Santa Clara, CA).
Autofluorescence Microscopy
The reactor effluents were examined by laser scanning confocal microscopy (Leica SP5 II, Leica Microsystems GmbH, Wetzlar, Germany) under the following conditions: excitation (405 nm laser) and emission (430-500 nm), which are used to visualize methanogens using their natural autofluorescent characteristics [23].
Transmission Electron Microscopy (TEM)
VLPs in the reactors were viewed using transmission electron microscope (TEM, Philips CM100 TEM, 100kV). TEM samples were prepared in general accordance with the methods described by Ackermann [24]. Samples were filtered through 0.2 µm low protein binding polyethersulfone (PES) syringe filters (Whatman, Clifton, NJ). Filtrate was placed in a 10 ml-polycarbonate tube (Seton Scientific, CA) and ultracentrifuged at 171,500×g and 4°C for 2 h (Beckman Coulter 70.1 Ti rotor, Fullerton, CA). The viral pellet was resuspended in 1 ml of 0.1 M ammonium acetate and centrifuged again at 79,302×g and 4°C for 1 h. The pellet was resuspended into 0.2 ml of 0.1 M ammonium acetate and stored at 4°C.
To prepare TEM grids, 3 μl of poly-L-lysine was placed on a 300 mesh carbon-stabilized formvar-coated copper grids (TedPella Inc, Reading, CA) for 1 min. Then the poly-L-lysine was removed by wicking using Whatman no. 1 filter paper. Viral suspension (5 μl) was placed on the grid for 2 min. Finally, 1μl of negative stain, phosphotungstic acid (PTA, 2%, pH 7.2) was added for 1 min, and the grid was wicked dry. Prepared grids were placed in a desiccator at least overnight until examination by TEM. The head diameter and tail length of VLPs were measured by ImageJ (version 1.43u). Micrographs were screened, and clear images of VLPs were selected for measurement of VLP size.
Epifluorescence Microscopy (EFM)
0.2 µm filtrate for EFM was collected daily for one week. Filtrate was harvested from both reactors: 0.5 h before and 4 h after the feeding of the daily-fed reactor (5 samples before and 5 samples after feeding from the daily-fed reactor); 3 samples before (0.5 h) and 4 samples after (0.5 h) the feeding from the hourly-fed reactor.
Nucleic acid stained VLPs were directly enumerated using Leica DM-LB microscope (Buffalo Grove, IL). VLPs were enumerated using EFM methods described by Noble and Fuhrman [25] with minor modification. Reactor samples were preserved in 10 mM sodium pyrophosphate, 2% formaldehyde and 94.5 mM of NaCl (final concentrations); preservation solutions used were prefiltered using a 0.02 µm Whatman Anotop™ syringe filter. Fixed solutions were held on ice for 15 min and filtered through Whatman 0.2 µm PES filters. Two µl of 1,000× SYBR Gold (Invitrogen Molecular Probes, Eugene, OR) was added to stain viral nucleic acid. After vortexing, samples were incubated on ice in the dark for at least 15 min. The stained VLPs were collected on 0.02 µm pore-sized AnodiscTM aluminum oxide-membrane filters (6809-6002, Whatman, Clifton, NJ). After drying the filter on a Kimwipe® tissue (Kimberly-Clark Corp., Roswell, GA) in the dark, the filter was placed on a glass slide and covered with a cover slip with freshly made mounting solution (50% glycerol and 50% PBS with 0.1% p- phenylenediamide). Slides were stored at -20°C until enumeration. For enumeration of VLPs, at least 10 images per filter were captured by digital camera (Leica DC 300F) using Image-Pro Plus 5.1 (Media Cybernetics, Inc., Bethesda, MD). Fields were randomly selected, avoiding the area near the supporting ring of the filter. Images were processed by IrfanView (version 4.27) (converted to grey images). VLPs were enumerated using ImageJ (version 1.43u).
Pulsed Field Gel Electrophoresis (PFGE)
Cellular materials in 1 L of sample from each reactor were initially separated from viruses by centrifuging at 3696×g and 4°C for 30 min in a Sorvall Legend RT tabletop centrifuge equipped with a swinging-bucket rotor (Cat. No. 75006441) (Thermo Scientific, Waltham, MA). Supernatant was collected and serially filtered through 0.45 and 0.20 µm pore size low protein binding polyethersulfone (PES) syringe filters (6780-2502 and 6780-2504, Whatman, Clifton, NJ). Viruses in the filtrates were concentrated using a series of centrifugal ultrafiltration units (Centricon UFC703008, Amicon Ultra-15 UFC903024 and Amicon Ultra-0.5 UFC503008; Millipore, Billerica, MA) following the manufacturer’s instructions. In addition, ultrafilter retentates were washed twice in SM buffer (100 mM NaCl, 8 mM MgSO4·7H2O, 50 mM Tris-Cl; without gelatin) and the final volume adjusted to 50 µl. The concentrate was treated with 1U of DNase I (AM2222, Life Technologies, Grand Island, NY) at 37°C for 1 h to degrade free DNA followed by 65°C for 15 min and the addition of 10 mM of EDTA (final concentration) to inactivate the DNase I.
Equal volumes of concentrate and 2% low-melting agarose (A9414, Sigma-Aldrich, St. Louis, MO) were mixed by gentle pipetting and transferred to plug molds. After solidification, plugs were incubated overnight in extraction buffer (100 mM EDTA, pH 8.0, 1% SDS and 1 mg/ml proteinase K (4333793, Life Technologies, Grand Island, NY)) at room temperature with gentle mixing on a rocker. The plugs were rinsed three times with 1X TE buffer for 30 min and stored at 4°C. Plugs and the DNA marker (N3551S, New England Biolabs, Ipswich, MA) were placed into wells of a 1% agarose gel (162-0137, BioRad, Hercules, CA) prepared in 0.5X TBE (wells were sealed with 1% agarose). PFGE was performed with CHEF-DR II System (BioRad, Hercules, CA) using the following conditions: 0.5X TBE, 6 V/cm, 15°C for 24 hours, switch times ramped from 1-25 seconds. The gel was stained with SYBR Green I (Molecular Probes, Eugene, OR) for 30 min and the image was acquired by a UVP GDS-8000 System (UVP, Upland, CA).
DNA Extraction
Genomic DNA from the reactors was isolated by UltraClean Soil DNA Isolation Kit (Mo Bio Laboratories, Carlsbad, CA). DNA extraction was performed following the manufacturers' protocol except the samples were disrupted using a FastPrep®-24 bead beater (MP Biomedicals, Solon, OH) at setting 5 m/s for 20 s.
Quantitative PCR (qPCR)
Quantification of 16S rRNA genes of Methanosaeta, Methanosarcina and Bacteria were performed using an Eppendorf Mastercycler® ep realplex and RealMasterMix kit (Eppendorf, Hauppauge, NY). Standards were prepared using PCR products of 16S rRNA genes of Methanosaeta concilii (DSM 6752), Methanosarcina barkeri (DSM 804) and Sphingopyxis TrD1 (GenBank® accession number: JN940802), which were cloned using TOPO TA Cloning Kit (Invitrogen Molecular Probes, Grand Island, NY). Plasmids were isolated by QIAprep Spin Miniprep Kit (Qiagen, Valencia, CA) and quantified using a NanoDrop 1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). Plasmids were linearized by restriction enzyme EcoRI (R6011, Promega Co., Madison, WI). Reaction mixes contained 2 μl DNA templates, 4.5 μl RealMasterMix and individual forward and reverse primers adjusted with water to 10 μl. Primers used for qPCR [26,27] are summarized in Table 2. For qPCR of Methanosaeta, thermal cycling conditions were as follows: initial denaturation at 95°C for 10 min, 50 cycles with denaturation for 10 s at 95°C, annealing for 20 s at 61.8°C, extension for 20 s at 68°C. The resulting standard curve for Methanosaeta had an efficiency of 98% (R2=0.9826) with a method detection limit of 1x104 16S rRNA gene copies per PCR reaction. For qPCR of Methanosarcina, thermal cycling conditions were: initial denaturation at 95°C for 10 min, 50 cycles with denaturation for 10 s at 95°C, annealing for 20 s at 64°C, extension for 20 s at 68°C. The resulting standard curve for Methanosarcina had an efficiency of 94% (R2=0.9982) with a method detection limit of 1x101 16SrRNA gene copies per PCR reaction. qPCR cycling conditions for bacteria were: initial denaturation at 95°C for 10 min, 50 cycles with denaturation for 15 s at 95°C, annealing for 30 s at 55.4°C, extension for 20 s at 68°C. Melting curves generated at the end of the qPCR reactions were routinely examined to verify the correct amplification The resulting standard curve for Bacteria had an efficiency of 84% (R2=0.9826) with a method detection limit of 3x103.
Table 2. Primers for Methanosaeta, Methanosarcina, Bacteria, and Archaea.
| Target Organisms | Primer | Sequence | Concentration | Reference |
|---|---|---|---|---|
| Primers used for qPCR | ||||
| Methanosaeta | MS1b | 5'-CCGGCCGGATAAGTCTCTTGA-3' | 0.5 μM | [27] |
| SAE835R | 5'-GACAACGGTCGCACCGTGGCC-3' | 0.5 μM | [27] | |
| Methanosarcina | MB1b | 5'-CGGTTTGGTCAGTCCTCCGG-3' | 0.3 μM | [27] |
| SAR835R | 5'-AGACACGGTCGCGCCATGCCT-3' | 0.3 μM | [27] | |
| Bacteria | 1114F | 5'-CGGCAACGAGCGCAACCC-3' | 0.5 μM | [26] |
| 1275R | 5'-CCATTGTAGCACGTGTGTAGCC-3' | 0.5 μM | [26] | |
| Primers used for cloning reactions | ||||
| Bacteria | 8F | 5'-AGAGTTTGATCCTGGCTCAG-3' | [28,29] | |
| Archaea | 21F | 5'-TACCTTGTTACGACTT-3' | [28,29] | |
| Universal | 1492R | 5'-TTCCGGTTGATCCYGCCGGA-3' | [28,29] | |
Clone Libraries and Phylogenetic Analysis
16S rRNA genes were amplified with forward primers specific for Bacteria, and Archaea combined with a Universal reverse primer [28,29] (Table 2). Clone libraries were constructed by using TOPO TA Cloning Kit (Invitrogen Molecular Probes, Grand Island, NY). 16S rRNA genes in clones were sequenced at the High Throughput Genomics Center at University of Washington from one end (Bacteria: 8F, Archaea: 21F or 1492R). Sequences were trimmed and assembled using Sequencher® (version 4.9, Gene Codes Corporation, Ann Arbor, MI), and sequences having similarity >99% were defined as an OTU (Operational Taxonomic Unit). Taxonomic affiliation of OTU was determined by RDP Classifier [30]. All sequences were deposited in the GenBank database under accession numbers KC961776 - KC961942.
Results
Morphology of VLPs
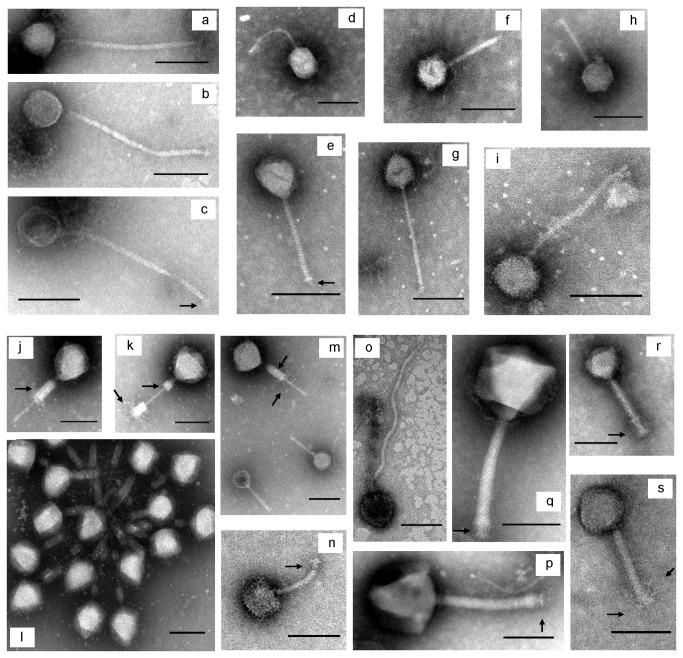
TEM images of VLPs from the reactors are shown in Figure 1. All of the observed VLPs were head-tailed morphologies, consistent with the descriptions of the order Caudovirales [31,32]. Within this morphology, two distinct variations were observed: isometric capsids with long noncontractile tails (characteristic of the family Siphoviridae; ex: Figure 1 a, b, c, e, g, i and o) in both reactors, and isometric capsids with long contractile tails (characteristic of the family Myoviridae; ex: Figure 1 j, p, q, r and s) only in the hourly reactor. One example of an additional morphology was observed (Figure 1d) that had a moderately elongated capsid. The only described family within the order Caudovirales not observed in this study was Podoviridae, which is characterized by short tails. Various facultative structures of VLPs were evident in the TEM micrographs (for a review of the description of facultative structures see 32). For example, tail sheaths (Figure 1 j, k and m), base plates (Figure 1 e, n and p), tail spikes (Figure 1e) and tail fibers (Figure 1 k, m, r, and s) were observed as indicated by arrows in the figures.
Figure 1. Selected transmission electron microscope (TEM) micrographs of virus-like particles (VLPs) in the reactors.
VLPs in the daily-fed reactor (a-i) and the hourly-fed reactor (j-s). VLPs morphologically similar to Siphoviridae (a, b, c, e, g, i and o) and to Myoviridae (j, p, q, r and s) were observed. Scale bar represents 100 nm.
Size Distribution of VLPs
The sizes of capsids and tails of 41 VLPs from the daily-fed reactor and 42 VLPs from the hourly-fed reactor were measured. Size distribution of VLPs varied between two reactors (Figure 2a) with smaller capsids and longer tails observed in the daily-fed reactor, and larger capsids and shorter tails in the hourly-fed reactor (Table 3).
Figure 2. Distribution of measured capsids diameters and tail lengths.
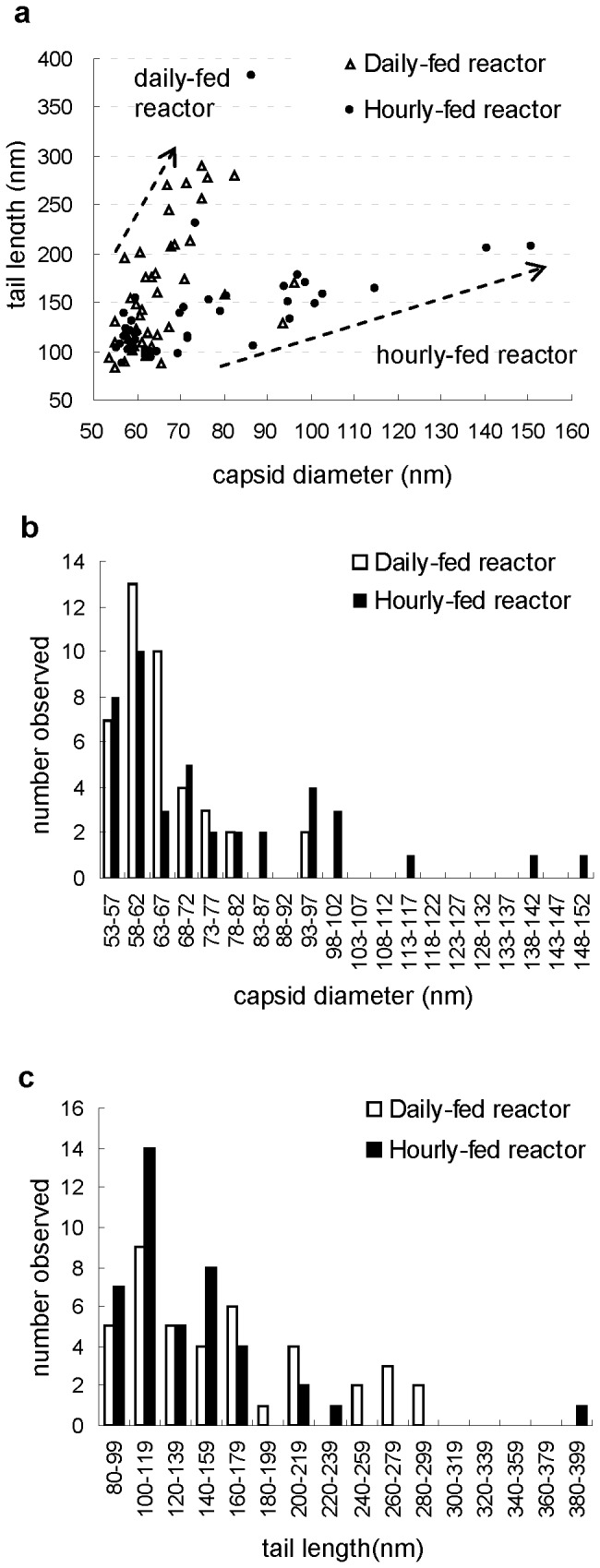
41 virus-like particles (VLPs) from the daily-fed reactor and 42 VLPs from the hourly-fed reactor were measured. a XY scatter chart, b size of capsids: X-axis shows capsid diameters and Y-axis shows number observed, c tail lengths: X-axis shows tail lengths and Y-axis shows number observed.
Table 3. Dimensions of VLPs observed in the reactors in comparison to previously documented phage families.
| reactor/phage families | Head diameter (nm) |
Tail length (nm) |
||||
|---|---|---|---|---|---|---|
| average | range | STD b | average | range | STD b | |
| daily-fed reactor | 66 | 54 - 96 | 9.6 | 162 | 84 - 289 | 61.4 |
| hourly-fed reactor | 76 | 55 - 151 | 22.6 | 137 | 87 - 382 | 51.4 |
| Siphoviridaea | 55 | 40 - 97 | -- | 191 | 79 - 593 | -- |
| Myoviridaea | 85 | 53 - 160 | -- | 167 | 80 - 485 | -- |
251 total phages examined (Ackermann 1998) [32]
STD, standard deviation
In the daily-fed reactor, the size distribution of isometric capsid diameters showed a distribution peak in the 58 nm - 62 nm group (Figure 2b) while no clear peak was found in the distribution of tail lengths (Figure 2c). Long flexible noncontractile tails were observed. Four of these ranged between 200 nm and 220 nm and 7 tails were longer than 240 nm.
In the hourly-fed reactor, the size distribution of head diameters also showed a peak in the 58 nm - 62 nm group (Figure 2b). VLPs with extremely large capsid were also observed (e.g. Figure 1p: 140 nm and 1q: 150 nm). Most tail lengths ranged from 80 nm to 240 nm with one exception of a VLP with a 380 nm tail (Figure 2c and Figure 1o). The distribution of tail lengths formed two peaks, one with tail length ranging from 100 nm to 119 nm and the other with tail length between 140 nm and 159 nm.
Concentration of VLPs using EFM
The average concentrations of VLPs in the reactors quantified using EFM are shown in Figure 3. Variation of VLP concentrations was low over the course of a week and before and after automated feeding. The concentration of VLPs were similar in the hourly-fed reactor (8.4 ± 4.3×107 VLPs/ml) than in the daily-fed reactor (7.1 ± 1.5×107 VLPs/ml). Variation shows the deviation among multiple samples collected from the reactors at different times, and no significant difference indicated between the two (T-test, P = 0.05).
Figure 3. Concentrations of Methanosaeta, Bacteria and virus-like particle (VLP) in the daily-fed and hourly-fed reactors.
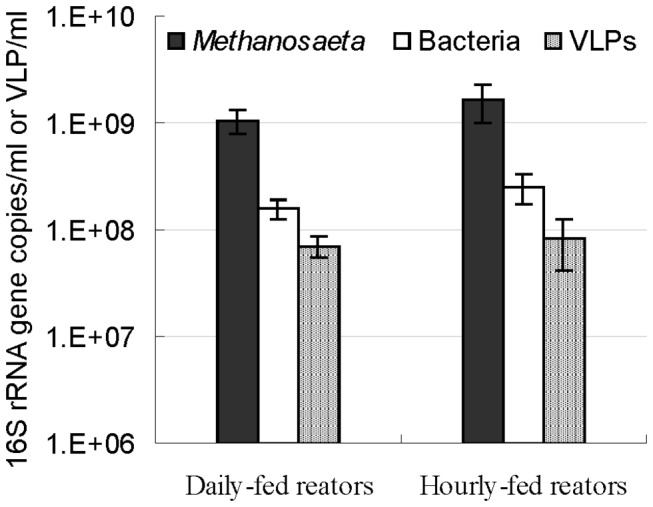
Values are the average of 6 samples for Methanosaeta and Bacteria, and 10 (daily-fed) and 7 (hourly-fed) samples for the VLP. Standard deviations were indicated by error bar.
Genome size of VLPs
Viral genome size distributions for the daily-fed and hourly-fed reactors are shown in the PFGE results (Figure 4). Different banding patterns were observed between the two reactors. Three bands (two major and one minor) were discovered in each reactor. The major (brightest) bands in the daily-fed and hourly-fed reactors were observed at approximately 80 and 85 kbp, respectively. The second brightest band was observed at approximately 35 kbp for both reactors. A third dim band in each reactor was observed at a higher molecular weight range (>240 kbp). Additional background smearing observed in the hourly-fed reactor, especially around 145 kbp may suggest DNA from multiple virus species with similar genome sizes.
Figure 4. Pulsed field gel electrophoresis (PFGE) of viral genomes from reactors.

Performance of the Acetate-fed Reactors
Both the daily-fed and hourly-fed reactors had neutral pH and consistent methane production prior to sampling. Monitoring of acetate concentrations, gas production and gas composition in the reactors showed that nearly all of the acetate was converted to methane (calculations data not shown). Neither reactor had any upsets or failure during the course of the study.
Dominant Reactor Microorganisms
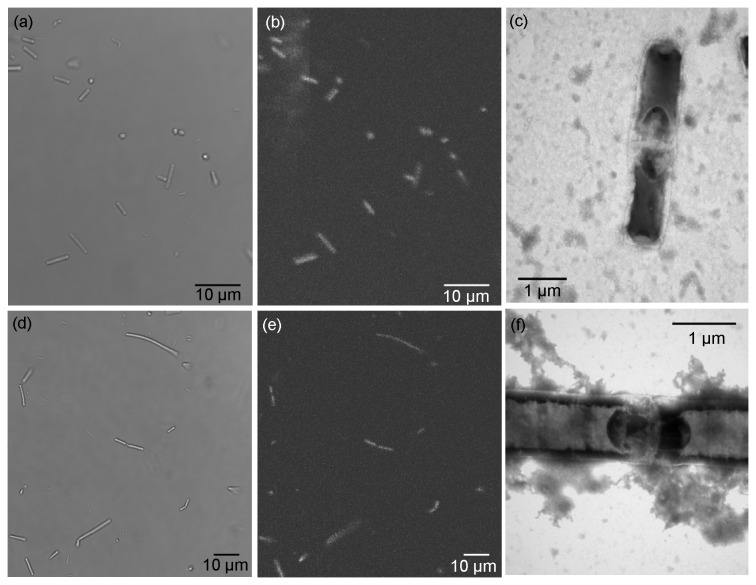
Three independent methods were used to demonstrate the dominance of Methanosaeta in the reactors including microscopy, qPCR and sequence recovery. Filamentous cells composed of several single flat-end rods (0.8-0.9 × 1.8-3.3 μm) were dominant in both reactors (Figure 5 a, c, d and f). These rods were non-motile and generally grew as long threads (3-50 μm). Autofluorescence characteristic of methanogenic Archaea was confirmed in the dominant microbial populations in the samples (Figure 5 b and e). The rod-shapes observed are consistent with Methanosaeta as described elsewhere [20,33,34]. Coccoidal morphotypes were observed with much lower frequency, some of which were autofluorescent.
Figure 5. Micrographs of enriched cultures in the daily-fed and hourly-fed reactors.
Micrographs (a-c) are from daily reactor and (d-f) are from hourly reactor. (a) and (d) are transmitted light bright field images. (b) and (e) are fluorescence micrographs. (c) and (f) are transmission electron microscope micrographs.
Methanosaeta qPCR signals dominated the reactors (Figure 3), while Methanosarcina was not detected (detection limit in the extract sample of 3x102 copies/ml of reactor effluent). Methanosaeta 16S rRNA gene copy concentrations were ~7× greater than total bacterial gene copy concentrations. Methanosaeta concentrations were not statistically different in the two reactors, while bacterial concentrations were higher in the hourly-fed reactor (T-test, P = 0.05).
Phylogenetic affiliation of archaeal and bacterial 16S rRNA genes from the two reactors as determined by clone sequencing are summarized in Table 4. In corroboration of the qPCR and microscopic evidence, the most frequently detected archaeal phylogenetic group was Methanosaeta in both reactors. One clone (out of 73 archaeal clones) in the daily-fed reactor aligned with the genus Methanosphaera (a Coccoidal-shaped microbe). No clone sequences from the group Methanosarcina were detected.
Table 4. Phylogenetic analysis of 16S rRNA gene in the daily-fed and hourly-fed reactors.
| Clone Library | Taxon a | No. of OTU b | No. of clone | Daily-fed (%) | Hourly-fed (%) |
|---|---|---|---|---|---|
| Archaea-21F | Methanosaeta | 4 | 36 | 96.4 | 100.0 |
| Methanosphaera | 1 | 1 | 3.6 | - | |
| Archaea-1492R | Methanosaeta | 4 | 36 | 100.0 | 100.0 |
| Bacteria | Proteobacteria | 6 | 43 | 47.8 | 44.7 |
| Firmicutes | 7 | 21 | 19.6 | 25.5 | |
| Bacteroidetes | 5 | 13 | 13.0 | 14.9 | |
| WWE1 | 2 | 7 | 2.2 | 12.8 | |
| minor groups c | 6 | 9 | 17.4 | 2.1 |
Taxon was determined by RDP classifier [30]; Archaea was grouped at the genus level and Bacteria was group at phylum level
OTU, Operational Taxonomic Unit
Clones with either 2 or less representatives in the library were from the groups Spirochaetes, Chloroflexi, Lentisphaerae, SR1 and unclassified bacteria.
For the bacterial domain, the most frequently observed clones in both reactors were from the phyla Proteobacteria, Firmicutes, and Bacteroidetes (Table 4). While other groups were consistent between the two reactors, more clones for the WWE1 (for a description of phylum WWE1 see 35) were obtained from the hourly-fed reactor. Of the Proteobacteria, the Arcobacter genus was represented by 35 clones, consisting of 45.7% of the Proteobacteria in the daily-fed and 29.8% in the hourly-fed reactors (data not shown).
Discussion
VLPs in Enriched Reactors
VLPs have been previously reported in anaerobic digesters [15,16]. However to the best of our knowledge, this is the first time that VLPs associated specifically with acetate-fed methanogenic consortia enriched in Methanosaeta have been characterized. Anaerobic digester sludge from municipal wastewater treatment plants have had higher reported VLP concentrations (e.g. 2.4×1010 VLPs/ml [16]) likely associated with higher microbial diversity in these systems compared to the study enrichment reactors. Still, the prevalence of Siphoviridae observed in the study reactors was consistent with Ackermann’s 2007 survey that showed this virus familyto be the most common tailed-phage documented in published electron micrograph images irrespective of the habitat. Siphovirdae were also the most abundant identified morphotype (16%) in a previously studied methanogenic digester treating wastewater from a beer brewery [15]. Observation of this common viral family is particularly interesting because although some archeoviruses have been reported to exhibit exceptionally complex morphotypes (e.g. linear, fusiforms, droplet and bottle shapes) [12,36], currently characterized viruses of methanogens typically have head-tail morphologies [37–39].
Several observations demonstrate that virus populations between two reactors differed. First, VLPs morphologically similar to Myoviridae were only found in the hourly-fed reactor (Figure 1). Second, different dominant VLP capsid sizes and tail lengths were observed in the two reactors (Figure 2a), a parameter thought to be highly conserved and uniform for individual virus species [32]. Third, PFGE banding patterns suggested that the viruses in the two reactors had different genome sizes (Figure 4, e.g. virus with 80 kbp genome in the daily-fed reactor and virus with 85 kbp in the hourly-fed reactor). Different virus populations may have developed in the reactors due to several contributing factors such as feeding schedule, HRT, re-inoculation of the daily-fed reactor, natural population drift in each reactor caused by unique “virus-host arm races”.
Enriched Cultures and Microbial Communities
One genus of the acetoclastic methanogens, Methanosaeta, was dominant in both reactors. Higher periodic acetate concentrations and shorter HRT in the daily-fed reactor was designed to favor Methanosarcina due to its higher growth rate (k) and lower acetate affinity (higher Ks) than Methanosaeta and Methanosarcina was historically dominant in the daily-fed reactor as previously reported [22]. However, dominance shifted from toward Methanosaeta by year 2007. Re-inoculation of the daily-fed reactor with sludge from municipal anaerobic digesters failed to reestablish Methanosarcina, as evidenced by qPCR, microscopic, and sequencing results presented in this work. The loss of Methanosarcina in the daily-fed reactor remains unexplained, but allowed comparison of VLP communities in two reactor systems that shared a dominant microbial population.
While the Bacteria were less prevalent than the Methanosaeta, their phylogeny still reveal interesting insights into the reactor microbial communities. The phyla Proteobacteria, Firmicutes and Bacteroidetes detected in the reactors have previously been reported as core microbial groups in several full-scale mesophilic anaerobic digesters [40] and in mesophilic bovine serum albumin-fed reactors [41]. One of the OTU detected in this study (containing 35 clones) within phylum Proteobacteria was assigned to genus Arcobacter and was most frequently detected in both the daily-fed (45.7%) and hourly-fed (29.8%) reactors (data not shown). The closest related sequence (NCBI accession number: GQ136513.1, 100% sequence coverage and similarity) to this OTU was a clone also found in an acetate enriched digester sample, implying that an anaerobic acetate enriched niche may be a favorable habit for Arcobacter. Sequences assigned to WWE1 were observed more frequently in the hourly-fed reactor, while the sequences affiliated with minor groups were more dominant in the daily-fed reactor, indicating different bacterial communities between the two reactors. Most of the bacterial 16S rRNA gene sequences recovered did not closely affiliate to any known isolated culture (similarity <97%) but were similar to the clones found in the anaerobic digesters [40–44], methanogenic consortium and landfill leachate [45] suggesting these anoxic systems may contain many bacterial species with yet unconfirmed metabolic roles.
VLPs and Enriched Cultures
Observations of persistent VLP populations in the reactors suggest that they are actively propagating in the system. Because viruses are obligate parasites, they only reproduce when their corresponding hosts (Bacteria or methanogenic Archaea in this study) are present. The feed for the reactors was prepared aseptically and evaluated by EFM to confirm that VLPs were not present. VLPs that entered the reactors with the anaerobic digestion sludge used to inoculate the reactors is not mathematically predicted to have persisted in the reactors due to the extended time since the last re-inoculation (>3 years prior to the study) in comparison to the systems’ retention times. Thus, replication of the observed VLPs was the most likely explanation for observation of VLP persistence in the system.
While replication of the virus population was supported by the data, the virus-to-bacteria ratio (VBR) values were lower in the study reactors than is often reported. The ratio of VLP concentrations to the prokaryotic cells (Methanosaeta and bacteria) concentration was 0.123 in the daily-fed reactor and 0.093 in the hourly-fed reactor (assuming 2 and 3.8 16S rRNA genes copies per organism for Methanosaeta [46] and Bacteria [47], respectively. In contrast to the study calculation, when VBR has been used to study relationships between viruses and bacteria in many natural aquatic ecosystems, the number of viruses are higher than the number of bacteria with typical VBRs ranging from 3 to 10 in aquatic ecosystems [48]. Assumptions used to estimate microorganism concentrations from 16S rRNA copy number may partially contribute to lower VBR values, particularly because the numbers of 16SrRNA genes in a chromosome can vary among species of the same genus and some methanogens have recently been reported to contain more than one chromosome [49] (though not yet studied for slow-growing Methanosaeta). However, lower VBR values similar to this study’s results have been reported in an oligotrophic lake (0.03-0.7) [50] and in rhizosphere soil (0.04) [51]. VBR values in Archaea-dominant environments have not been reported.
Finding of low VLPs concentration in the two Methanosaeta-dominant reactors studied here may due to several reasons. First, viruses in reactors might prefer lysogenic or pseudolysogenic [52] life cycles, in which viruses persist inside host cells and thus were not counted using the study methods. Second, it has been shown that the VBR is higher in nutrient-rich, more productive environments [48]. However, the acetate-fed methanogenic reactors in this study are not classified as productive environments for viruses. Under anaerobic conditions, limited energy is gained by slow growing Methanosaeta when acetate is converted to methane, and only limited substrate (e.g. vitamins) was available for the growth of bacteria (although metabolic roles of bacteria in reactors is unknown). Third, a positive correlation between VBR and host community diversity has been previously observed [53], suggesting that the VBR may be an indicator of host community diversity. For the reactors in the current study, Methanosaeta was found to be dominant in the daily-fed and hourly-fed reactors and low diversity of microbial communities was observed, which may explain with the low VBR observed in two enriched reactors.
Despite persistent VLP presence in both Methanosaeta dominant reactors, upset or failure of these methanogenic reactors was not observed over the study period. This might be explained by the presence of temperate viruses, chronic infections or co-evolution of virus and host populations in reactors. Limited energy conditions and a slow growing host under anaerobic conditions may be favorable for lysogenic or pseudolysogenic life cycles. Additionally, some crenarchaeal viruses have been found to be chronically produced without host cells lysis [12,54,55]. Although viruses of acetoclastic methanogens have not yet been reported, this hypothesis seems less likely because currently identified head-tail morphotypes, such as those observed in this study, have previously been reported as lytic [37,39]. Finally, an evolutionary arms race between viruses and hosts could result in minor changes in population structure without the changing of community stability [56]. Nevertheless, this theory requires more investigation for methanogenic habitats.
Viral-host dynamics for the archaea are complex. Further understanding of the viral life strategies (e.g. lysis, lysogeny, and pseudolysogeny) and coevolution with host will lead to improved understanding of the impact of acetoclastic archeoviruses on microbial ecology of anaerobic digesters.
Acknowledgments
We would like to acknowledge Jody Deming’s research group for generous assistance with epifluorescence microscopy.
Funding Statement
This material is based upon work supported by the National Science Foundation under grant number CBE-0930877. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Grosskopf R, Janssen PH, Liesack W (1998) Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl Environ Microbiol 64: 960-969. PubMed: 9501436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Scholten JCM, Stams AJM (2000) Isolation and characterization of acetate-utilizing anaerobes from a freshwater sediment. Microb Ecol 40: 292-299. PubMed: 12035087. [DOI] [PubMed] [Google Scholar]
- 3. Elberson MA, Sowers KR (1997) Isolation of an aceticlastic strain of Methanosarcina siciliae from marine canyon sediments and emendation of the species description for Methanosarcina siciliae . Int J Syst Bacteriol 47: 1258-1261. doi: 10.1099/00207713-47-4-1258. PubMed: 9336940. [DOI] [PubMed] [Google Scholar]
- 4. Steinberg LM, Regan JM (2008) Phylogenetic Comparison of the Methanogenic Communities from an Acidic, Oligotrophic Fen and an Anaerobic Digester Treating Municipal Wastewater Sludge. Appl Environ Microbiol 74: 6663-6671. doi: 10.1128/AEM.00553-08. PubMed: 18776026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Smith KS, Ingram-Smith C (2007) Methanosaeta, the forgotten methanogen? Trends Microbiol 15: 150-155. doi: 10.1016/j.tim.2007.02.002. PubMed: 17320399. [DOI] [PubMed] [Google Scholar]
- 6. Conrad R (1999) Contribution of hydrogen to methane production and control of hydrogen concentrations in methanogenic soils and sediments. FEMS Microbiol Ecol 28: 193-202. doi: 10.1111/j.1574-6941.1999.tb00575.x. [DOI] [Google Scholar]
- 7. McCarty PL (2001) The development of anaerobic treatment and its future. Water Sci Technol 44: 149-156. PubMed: 11730130. [PubMed] [Google Scholar]
- 8. Chen Y, Cheng JJ, Creamer KS (2008) Inhibition of anaerobic digestion process: A review. Bioresour Technol 99: 4044-4064. doi: 10.1016/j.biortech.2007.01.057. PubMed: 17399981. [DOI] [PubMed] [Google Scholar]
- 9. Nachaiyasit S, Stuckey DC (1997) The effect of shock loads on the performance of an anaerobic baffled reactor (ABR) .1. Step changes in feed concentration at constant retention time. Water Res 31: 2737-2746. doi: 10.1016/S0043-1354(97)00133-4. [DOI] [Google Scholar]
- 10. Kim M, Ahn YH, Speece RE (2002) Comparative process stability and efficiency of anaerobic digestion; mesophilic vs. thermophilic. Water Res 36: 4369-4385. doi: 10.1016/S0043-1354(02)00147-1. PubMed: 12420941. [DOI] [PubMed] [Google Scholar]
- 11. Kroeker EJ, Schulte DD, Sparling AB, Lapp HM (1979) Anaerobic treatment process stability. J Wat Pollut Control Fed; 51: 718-727 [Google Scholar]
- 12. Pina M, Bize A, Forterre P, Prangishvili D (2011) The archeoviruses. FEMS Microbiol Rev 35: 1035-1054. doi: 10.1111/j.1574-6976.2011.00280.x. PubMed: 21569059. [DOI] [PubMed] [Google Scholar]
- 13. Suttle CA (2007) Marine viruses - major players in the global ecosystem. Nat Rev Microbiol 5: 801-812. doi: 10.1038/nrmicro1750. PubMed: 17853907. [DOI] [PubMed] [Google Scholar]
- 14. Thingstad TF (2000) Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol Oceanogr 45: 1320-1328. doi: 10.4319/lo.2000.45.6.1320. [DOI] [Google Scholar]
- 15. Park MO, Ikenaga H, Watanabe K (2007) Phage diversity in a methanogenic digester. Microb Ecol 53: 98-103. doi: 10.1007/s00248-006-9053-9. PubMed: 17186158. [DOI] [PubMed] [Google Scholar]
- 16. Wu QL, Liu WT (2009) Determination of Virus abundance, diversity and distribution in a municipal wastewater treatment plant. Water Res 43: 1101-1109. doi: 10.1016/j.watres.2008.11.039. PubMed: 19095276. [DOI] [PubMed] [Google Scholar]
- 17. Jeris JS, McCarty PL (1965) The Biochemistry of Methane Fermentation Using C14 Tracers. J Wat Pollut Control Fed; 37: 178-192 [Google Scholar]
- 18. Smith PH, Mah RA (1966) Kinetics of Acetate Metabolism during Sludge. Digestion - Appl Microbiol 14: 368-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Leclerc M, Delgènes JP, Godon JJ (2004) Diversity of the archaeal community in 44 anaerobic digesters as determined by single strand conformation polymorphism analysis and 16S rDNA sequencing. Environ Microbiol 6: 809-819. doi: 10.1111/j.1462-2920.2004.00616.x. PubMed: 15250883. [DOI] [PubMed] [Google Scholar]
- 20. Janssen PH (2003) Selective enrichment and purification of cultures of Methanosaeta spp. J Microbiol Methods 52: 239-244. doi: 10.1099/jmm.0.05045-0. PubMed: 12459244. [DOI] [PubMed] [Google Scholar]
- 21. Carbonero F, Oakley BB, Purdy KJ (2010) Improving the isolation of anaerobes on solid media: the example of the fastidious Methanosaeta . J Microbiol Methods 80: 203-205. doi: 10.1016/j.mimet.2009.11.013. PubMed: 19969029. [DOI] [PubMed] [Google Scholar]
- 22. Conklin A, Stensel HD, Ferguson J (2006) Growth kinetics and competition between Methanosarcina and Methanosaeta in mesophilic anaerobic digestion. Water Environ Res 78: 486-496. doi: 10.2175/106143006X95393. PubMed: 16752610. [DOI] [PubMed] [Google Scholar]
- 23. Doddema HJ, Vogels GD (1978) Improved identification of methanogenic bacteria by fluorescence microscopy. Appl Environ Microbiol 36: 752-754. PubMed: 103504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ackermann HW (2009) Basic phage electron microscopy. Methods Mol Biol 501: 113-126. doi: 10.1007/978-1-60327-164-6_12. PubMed: 19066816. [DOI] [PubMed] [Google Scholar]
- 25. Noble RT, Fuhrman JA (1998) Use of SYBR Green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat Microb Ecol 14: 113-118. doi: 10.3354/ame014113. [DOI] [Google Scholar]
- 26. Denman SE, McSweeney CS (2006) Development of a real-time PCR assay for monitoring anaerobic fungal and cellulolytic bacterial populations within the rumen. FEMS Microbiol Ecol 58: 572-582. doi: 10.1111/j.1574-6941.2006.00190.x. PubMed: 17117998. [DOI] [PubMed] [Google Scholar]
- 27. Shigematsu T, Tang Y, Kawaguchi H, Ninomiya K, Kijima J et al. (2003) Effect of dilution rate on structure of a mesophilic acetate-degrading methanogenic community during continuous cultivation. J Biosci Bioeng 96: 547-558. doi: 10.1016/S1389-1723(04)70148-6. PubMed: 16233572. [DOI] [PubMed] [Google Scholar]
- 28. DeLong EF (1992) Archaea in coastal marine environments. Proc Natl Acad Sci U S A 89: 5685-5689. doi: 10.1073/pnas.89.12.5685. PubMed: 1608980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lane DJ (1991) 16S/23S rRNA sequencing. in Nucleic acid techniques in bacterial systematics. J. eds Stackebrandt E., Goodfellow M. (Wiley & Sons, Chichester, United Kingdom) . 115–175 [Google Scholar]
- 30. Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261-5267. doi: 10.1128/AEM.00062-07. PubMed: 17586664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ackermann HW (2007) 5500 Phages examined in the electron microscope. Arch Virol 152: 227-243. doi: 10.1007/s00705-006-0849-1. PubMed: 17051420. [DOI] [PubMed] [Google Scholar]
- 32. Ackermann HW (1998) Tailed bacteriophages: The order Caudovirales . Adv Virus Res 51: 135-201. doi: 10.1016/S0065-3527(08)60785-X. PubMed: 9891587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mizukami S, Takeda K, Akada S, Fujita T (2006) Isolation and characteristics of Methanosaeta in paddy field soils. Biosci Biotechnol Biochem 70: 828-835. doi: 10.1271/bbb.70.828. PubMed: 16636448. [DOI] [PubMed] [Google Scholar]
- 34. Zehnder AJ, Huser BA, Brock TD, Wuhrmann K (1980) Characterization of an acetate-decarboxylating, non-hydrogen-oxidizing methane bacterium. Arch Microbiol 124: 1-11. doi: 10.1007/BF00407022. PubMed: 6769415. [DOI] [PubMed] [Google Scholar]
- 35. Chouari R, Le Paslier D, Dauga C, Daegelen P, Weissenbach J et al. (2005) Novel major bacterial candidate division within a municipal anaerobic sludge digester. Appl Environ Microbiol 71: 2145-2153. doi: 10.1128/AEM.71.4.2145-2153.2005. PubMed: 15812049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Prangishvili D, Forterre P, Garrett RA (2006) Viruses of the Archaea: a unifying view. Nat Rev Microbiol 4: 837-848. doi: 10.1038/nrmicro1527. PubMed: 17041631. [DOI] [PubMed] [Google Scholar]
- 37. Meile L, Jenal U, Studer D, Jordan M, Leisinger T (1989) Characterization of psiM1, a virulent phage of Methanobacterium thermoautotrophicum Marburg. Arch Microbiol 152: 105-110. doi: 10.1007/BF00456085. [DOI] [Google Scholar]
- 38. Jordan M, Meile L, Leisinger T (1989) Organization of Methanobacterium thermoautotrophicum bacteriophage psi M1. DNA - Mol Gen Genet 220: 161-164. [DOI] [PubMed] [Google Scholar]
- 39. Nolling J, Groffen A, Devos WM (1993) phiF1 and phiF3, 2 novel virulent, archaeal phages infecting different thermophilic strains of the genus Methanobacterium . J Gen Microbiol 139: 2511-2516. doi: 10.1099/00221287-139-10-2511. [DOI] [Google Scholar]
- 40. Rivière D, Desvignes V, Pelletier E, Chaussonnerie S, Guermazi S et al. (2009) Towards the definition of a core of microorganisms involved in anaerobic digestion of sludge. ISME J 3: 700-714. doi: 10.1038/ismej.2009.2. PubMed: 19242531. [DOI] [PubMed] [Google Scholar]
- 41. Tang Y, Shigematsu T, Morimura S, Kida K (2005) Microbial community analysis of mesophilic anaerobic protein degradation process using bovine serum albumin (BSA)-fed continuous cultivation. J Biosci Bioeng 99: 150-164. doi: 10.1263/jbb.99.150. PubMed: 16233772. [DOI] [PubMed] [Google Scholar]
- 42. Ito T, Yoshiguchi K, Ariesyady HD, Okabe S (2011) Identification of a novel acetate-utilizing bacterium belonging to Synergistes group 4 in anaerobic digester sludge. ISME J 5: 1844-1856. doi: 10.1038/ismej.2011.59. PubMed: 21562600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shigematsu T, Tang Y, Mizuno Y, Kawaguchi H, Morimura S et al. (2006) Microbial diversity of mesophilic methanogenic consortium that can degrade long-chain fatty acids in chemostat cultivation. J Biosci Bioeng 102: 535-544. doi: 10.1263/jbb.102.535. PubMed: 17270719. [DOI] [PubMed] [Google Scholar]
- 44. Krakat N, Schmidt S, Scherer P (2011) Potential impact of process parameters upon the bacterial diversity in the mesophilic anaerobic digestion of beet silage. Bioresour Technol 102: 5692-5701. doi: 10.1016/j.biortech.2011.02.108. PubMed: 21435870. [DOI] [PubMed] [Google Scholar]
- 45. Limam RD, Bouchez T, Chouari R, Li T, Barkallah I et al. (2010) Detection of WWE2-related Lentisphaerae by 16S rRNA gene sequencing and fluorescence in situ hybridization in landfill leachate. Can J Microbiol 56: 846-852. doi: 10.1139/W10-065. PubMed: 20962908. [DOI] [PubMed] [Google Scholar]
- 46. Barber RD, Zhang L, Harnack M, Olson MV, Kaul R et al. (2011) Complete genome sequence of Methanosaeta concilii, a specialist in aceticlastic methanogenesis. J Bacteriol 193: 3668-3669. doi: 10.1128/JB.05031-11. PubMed: 21571998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fogel GB, Collins CR, Li J, Brunk CF (1999) Prokaryotic Genome Size and SSU rDNA Copy Number: Estimation of Microbial Relative Abundance from a Mixed. Population - Microb Ecol 38: 93-113. doi: 10.1007/s002489900162. [DOI] [PubMed] [Google Scholar]
- 48. Wommack KE, Colwell RR (2000) Virioplankton: viruses in aquatic ecosystems. Microbiol Mol Biol Rev 64: 69-114. doi: 10.1128/MMBR.64.1.69-114.2000. PubMed: 10704475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hildenbrand C, Stock T, Lange C, Rother M, Soppa J (2011) Genome Copy Numbers and Gene Conversion in Methanogenic. Archaea - J Bacteriol 193: 734-743. doi: 10.1128/JB.01016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tapper MA, Hicks RE (1998) Temperate viruses and lysogeny in Lake Superior bacterioplankton. Limnol Oceanogr 43: 95-103. doi: 10.4319/lo.1998.43.1.0095. [DOI] [Google Scholar]
- 51. Ashelford KE, Day MJ, Fry JC (2003) Elevated abundance of bacteriophage infecting bacteria in soil. Appl Environ Microbiol 69: 285-289. doi: 10.1128/AEM.69.1.285-289.2003. PubMed: 12514006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Łoś M, Węgrzyn G (2012) Pseudolysogeny. Adv Virus Res 82: 339-349. doi: 10.1016/B978-0-12-394621-8.00019-4. PubMed: 22420857. [DOI] [PubMed] [Google Scholar]
- 53. Tuomi P, Fagerbakke KM, Bratbak G, Heldal M (1995) Nutritional enrichment of a microbial community: The effects on activity, elemental composition, community structure and virus production. FEMS Microbiol Ecol 16: 123-134. doi: 10.1111/j.1574-6941.1995.tb00276.x. [DOI] [Google Scholar]
- 54. Prangishvili D, Vestergaard G, Häring M, Aramayo R, Basta T et al. (2006) Structural and genomic properties of the hyperthermophilic archaeal virus ATV with an extracellular stage of the reproductive cycle. J Mol Biol 359: 1203-1216. doi: 10.1016/j.jmb.2006.04.027. PubMed: 16677670. [DOI] [PubMed] [Google Scholar]
- 55. Prangishvili D, Garrett RA (2005) Viruses of hyperthermophilic Crenarchaea. Trends Microbiol 13: 535-542. doi: 10.1016/j.tim.2005.08.013. PubMed: 16154357. [DOI] [PubMed] [Google Scholar]
- 56. Andersson AF, Banfield JF (2008) Virus population dynamics and acquired virus resistance in natural microbial communities. Science 320: 1047-1050. doi: 10.1126/science.1157358. PubMed: 18497291. [DOI] [PubMed] [Google Scholar]