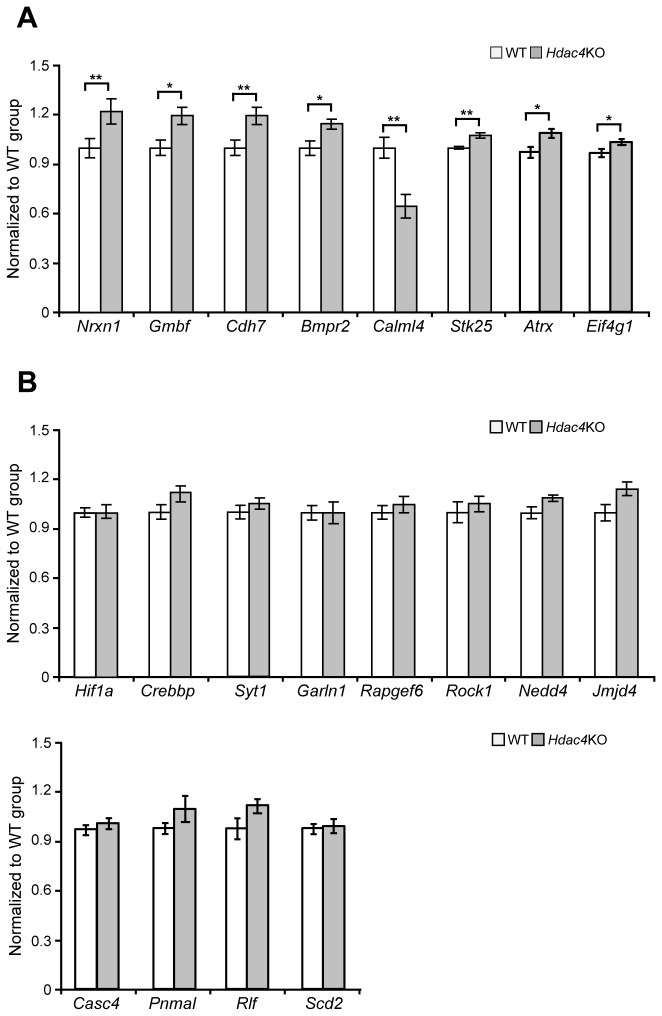
Figure 3. Validation of the genes predicted to be downregulated in Hdac4 knock-out P3 brains.
(A) Nrxn1 (neurexin1), Gmbf (glia maturation factor beta), Cdh7 (cadherin 7), Bmpr2 (bone morphogenic protein receptor, type II), Stk25 (serine/threonine kinase 25), Atrx (alpha thalassemia/mental retardation syndrome X-linked homolog) and Eif4g1 (eukaryotic translation initiation factor 4, gamma 1) were modestly upregulated in Hdac4KO P3 brains. Calml4 (calmodulin-like 4) was the only gene that was downregulated (B) The expression level of Hif1α (hypoxia inducible factor 1 alpha subunit), Crebbp (CREB-binding protein), Syt1 (synaptotagemin 1) Garln1 (GTPase activating RANGAP domain like 1 protein), Rapgef6 (Rap guanine nucleotide exchange factor GEF6), Rock1 (Rho-associated coiled-coil containing protein kinase 1), Nedd4 (NEDD4 binding protein), Jmjd4 (jumonji containing protein 4), Casc4 (cancer susceptibility candidate 4), Pnmal (PNMA-like 2), Rlf (rearranged L-myc fusion sequence) and Scd2 (stearoyl-Coenzyme A desaturase 2) did not change between Hdac4KO and WT P3 brains. All mRNA expression levels were assessed by Taqman qPCR and presented as a relative expression ratio to the geometric mean of three housekeeping genes Atp5b, Canx, Rpl13α. Error bars are S.E.M (n>7). *p<0.05, **p<0.01.