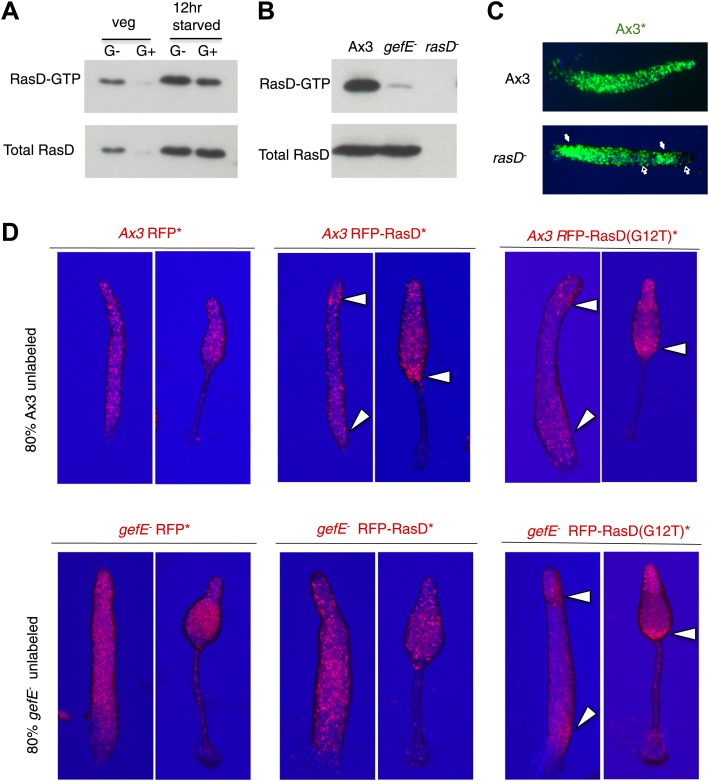
Figure 6. GefE activates RasD.
(A) Comparison of the levels of activated RasD-GTP and total RasD by Western blot in vegetative or 12 hr starved cells grown in the presence or absence of glucose. (B) Comparison of activated RasD-GTP and total RasD levels by Western blot in wild type Ax3, gefE− or rasD− cells. (C) GFP-labelled (*) Ax3 wild type cells mixed at 10:90 ratio with unlabelled Ax3 or rasD− mutant cells. Closed arrows show enrichment of wild type cells in pstO and pstB populations. Open arrows show reciprocal enrichment of rasD− cells. (D) RasD overexpression results in GefE dependent bias towards the pstO and pstB cell fates. 20% cells constitutively expressing RFP, RFP-RasD or RFP-RasD(G12T) were mixed with 80% unlabelled parental cells. When wild type Ax3 cells overexpress RFP-RasD or RFP-RasD(G12T) they become enriched in the pstO and pstB populations (arrows). Only gefE− cells that express constitutively activated RasD(G12T) are enriched in pstO and pstB populations (arrows).
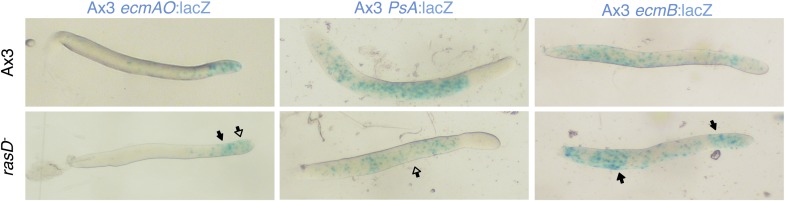
Figure 6—figure supplement 1. rasD− mutant cells avoid pstO and pstB fates in chimera with wild type cells.