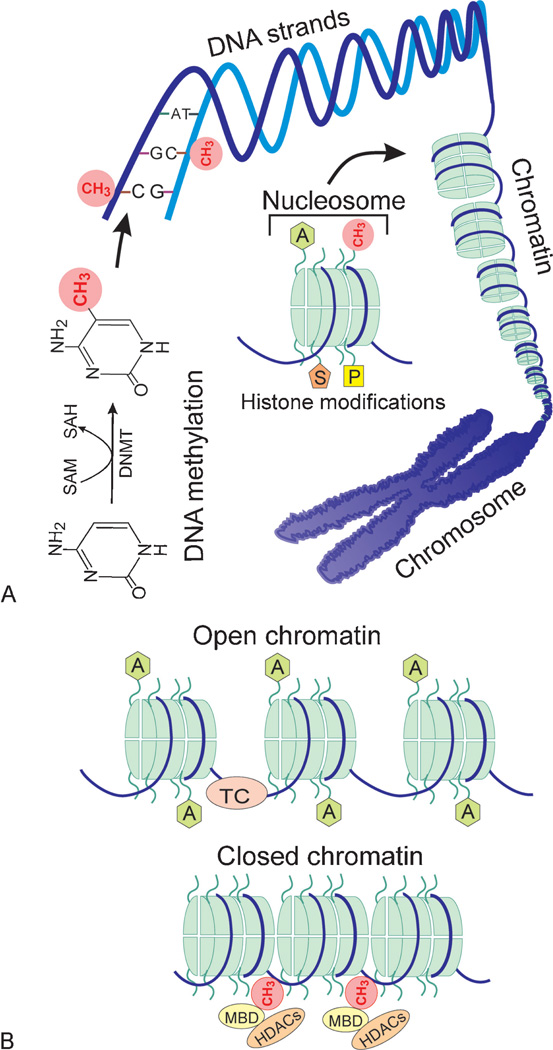
Figure 1.
Epigenetic gene regulation. (A) The overall organization of DNA in relation to epigenetic modifications. DNA methylation is mediated through DNA methyltransferases (DNMTs) by transferring a methyl group from S-adenosylmethionine (SAM) to the carbon-5 position of a cytosine base (shown as a red circle). DNA methylation typically occurs on cytosines (C) preceding guanines (G) at both strands of the double helix. DNA strands are wrapped around histone octamers to form nucleosomes that are building blocks of chromatin. The chromatin in turn is organized into a chromosome. The specific structure of nucleosomes facilitates epigenetic gene regulation. Histone tails protruding from each nucleosome can undergo numerous posttranslational modifications such as acetylation (A in a green hexagon), methylation (CH3 in red circles), sumoylation (S in a brown pentagon), and phosphorylation (P in a yellow rectangle). Such modifications along with DNA methylation can influence chromatin remodeling. (B) The role of chromatin remodeling in gene expression. The open and closed structure of chromatin is regulated by interactions of DNA methylation, numerous histone modifications, and noncoding RNAs. Only shown are DNA methylation (CH3 in red circles) and acetylation of histone tails (A in green hexagons) for a simplified view. Typically, demethylation of CpG dinucleotides (absence of red circles) and acetylation of histone tails (green hexagons) result in an open chromatin structure, which allows binding of the transcription complex (TC as a pink ellipse) to a specific sequence leading to transcription. In contrast, upon methylation of cytosine residues (red circles) by DNA methyltransferases, methyl binding domain proteins (MBD as yellow ellipses) bind to the methylated DNA and recruit histone deacetylases (HDACs as brown ellipses). These events induce deacetylation of histone tails, compaction of the chromatin, and thus transcriptional silencing due to inability of the TC to bind to DNA.