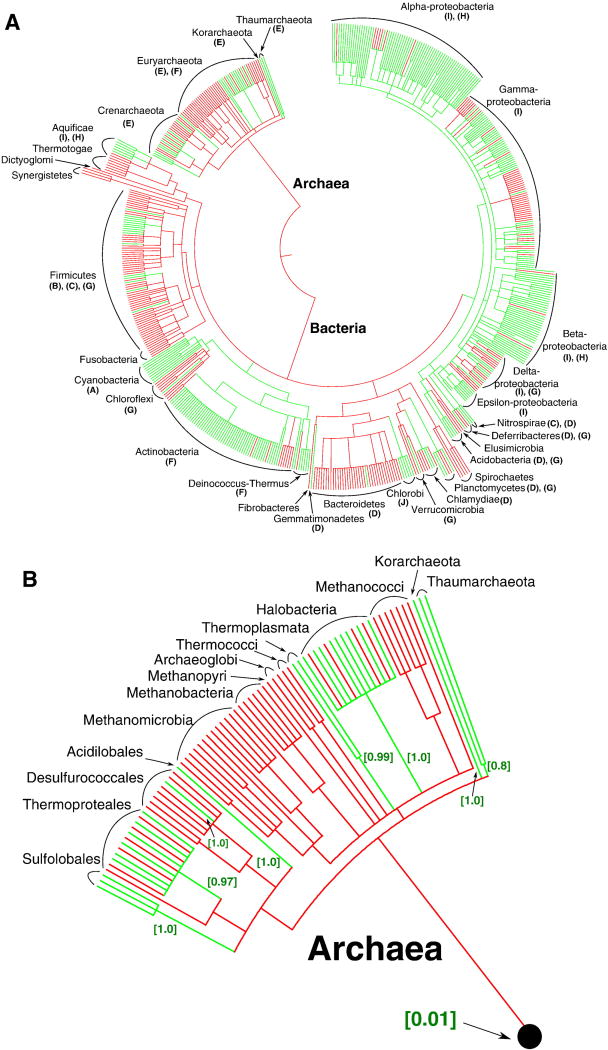
Figure 2.
Presence and absence of cytochrome b (COG1290), used as a marker of the cytochrome bc1 complexes, mapped on the ribosomal protein-based phylogenetic tree of prokaryotes [233, 234]. Branch lengths do not exactly reflect the evolutionary distance between the nodes. The assignment of proteins from the RefSeq release 45 (Jan 07, 2011) to the Clusters of Orthologous Groups (COGs) [236] was taken from the NCBI FTP site (ftp://ftp.ncbi.nih.gov/pub/wolf/COGs/Prok1202/). The redundancy in the list of complete genomes from the RefSeq release 45 was reduced by manually removing species of the same genera, which resulted in a list of 582 prokaryotic species. Taxonomy data from the NCBI (http://www.ncbi.nlm.nih.gov/taxonomy) [235] to the level of family were used to map the taxonomy for these genomes on the aforementioned large-scale tree. For calculations, the set of 582 bacterial genomes was further reduced to a compact set of 102 bacterial genomes. Within bacteria, we selected genomes which contained cytochromes b (68 genomes), as well as genomes of closely related species which did not contain cytochrome b. In calculations with the COUNT software, a full sample of 115 archaeal genomes and a compact set of 102 bacterial genomes from all major phyla were used, which resulted in a sample of 217 genomes. 29 COGs which occur in at least half of major bacterial and archaeal phyla and do not contain more than 10 members in each genome were selected randomly. These COGs were used as a reference for the estimation of typical rates of gene losses, gene gains and other parameters in COUNT by the “Gain-Loss-Duplication” model with default parameters. For the reference COGs and the cytochrome b COG1290, respectively, the occurrences in each of 217 sampled genomes were calculated as described above. The rates of gene losses and gains were optimized on a subset of 217 genomes chosen to satisfy the computational requirements of the program.
(A) Phylogenetic tree of prokaryotes. For the phyla that contain cytochromes b, the letters in brackets indicate the clades in Figure 1 that include cytochrome b sequences found in these phyla.
(B) Enlarged archaeal clade. The estimated probabilities of independent acquisition of cytochrome(s) b in each group are given in square brackets.