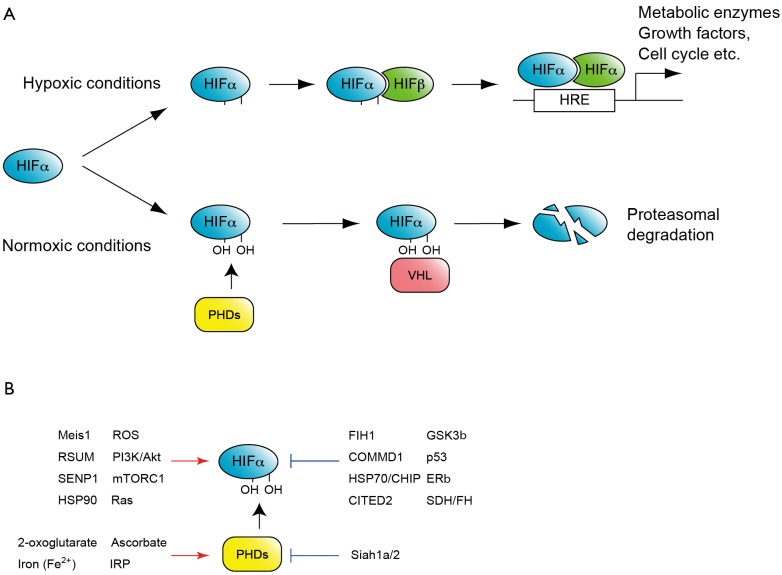
Figure 1.
O2 dependent HIF regulation. A. One of the classic and well-studied mechanisms regulating HIF activity by O2. Under normoxic conditions HIFα is hydroxylated by prolyl hydroxylase domain-containing enzymes (PHDs), and recognized by the E3-ubiquitin-ligase, von Hippel-Lindau (VHL) to be subjected to proteasome-mediated degradation. Under hypoxic conditions HIFα is stable and together with constitutive HIFα, binds to hypoxia response element (HRE) to activate hundreds of target genes; B. Factors activate or repress HIF activity. Note that these factors modulate HIF at several different levels. For example Meis1 activates the transcription of HIFα mRNA, whereas RSUME and SEMP1 mediate SUMOylation of HIFα proteins