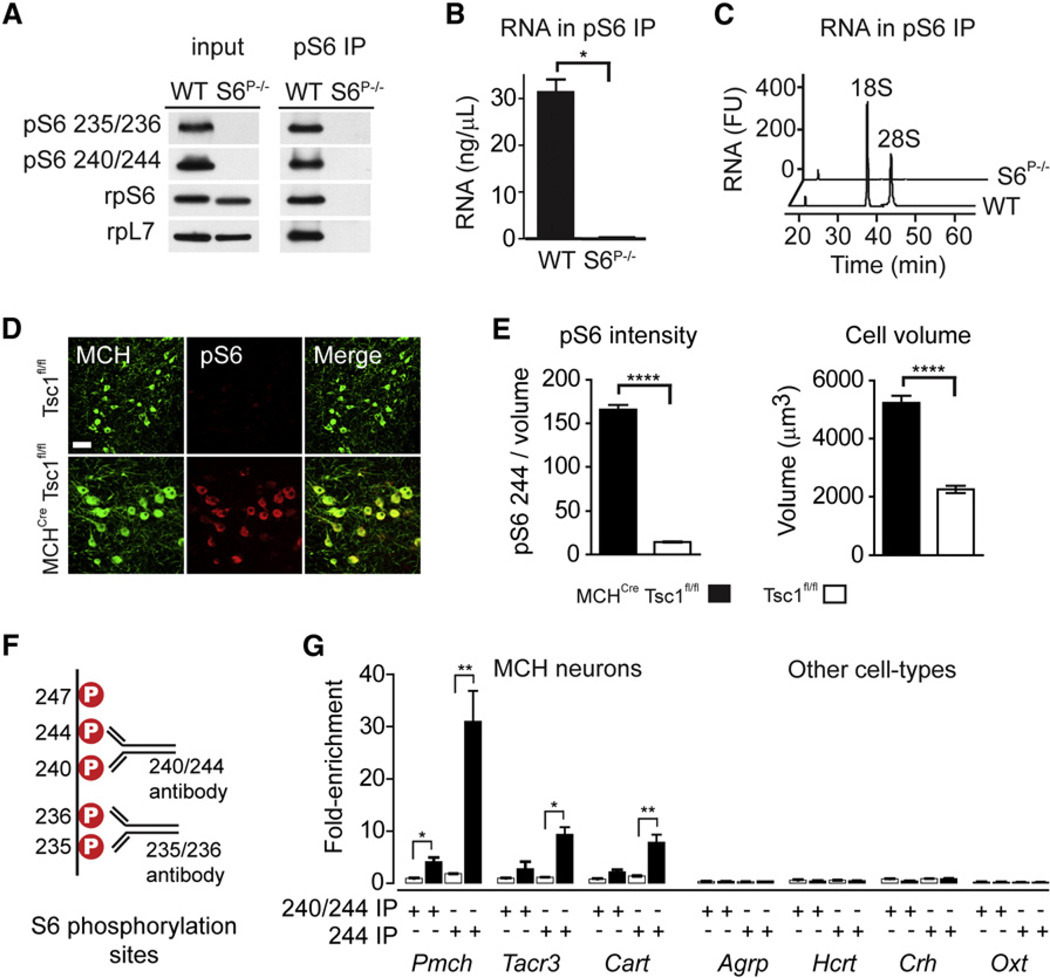
Figure 3. Selective Capture of Phosphorylated Ribosomes In Vitro and In Vivo.
(A) Western blot for ribosomal proteins from wild-type or S6S5A MEFs. Input lysate (left) and the pS6 240/244 immunoprecipitate (right) are shown.
(B) RNA yield from pS6 240/244 immunoprecipitates from wild-type and S6S5A MEFs.
(C) Bioanalyzer analysis of immunoprecipitated RNA from wild-type and S6S5A MEFs. 18S and 28S ribosomal RNA are labeled. FU, fluorescence units.
(D) Colocalization of MCH and pS6 in the hypo-thalamus of Tsc1fl/fl and MCHCre Tsc1fl/fl mice. Scale bar, 50 mm.
(E) Left: pS6 244 immunostaining density in MCH neurons following Tsc1 deletion. Right: volume of MCH neurons following Tsc1 deletion.
(F) Five major S6 phosphorylation sites and the diphospho-motifs recognized by commonly used pS6 antibodies.
(G) Enrichment of a panel of cell-type-specific genes following immunoprecipitation with antibodies recognizing pS6 240/244 or only pS6 244. Black bars represent immunoprecipitates from hypothalamic homogenates of MCHCre Tsc1fl/fl mice, whereas white bars represent Tsc1fl/fl controls. The enrichment of cell-type-specific genes in pS6 immunoprecipitates was determined by Taqman.
*p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 using two-tailed unpaired t test. All error bars are mean ±SEM. See also Figure S2.