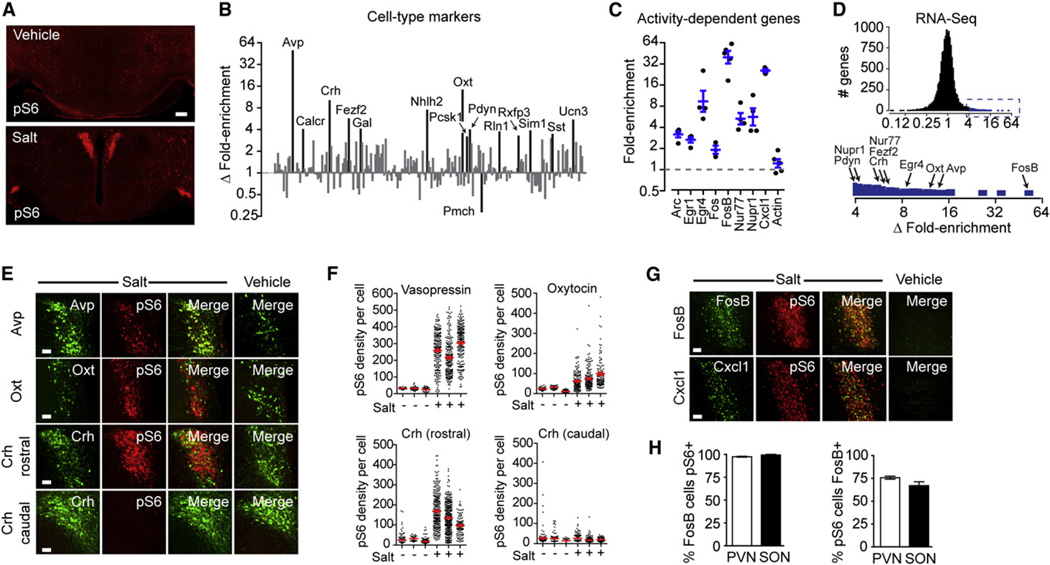
Figure 4. Identification of Neurons Activated by Salt Challenge.
(A) Hypothalamic staining for pS6 244 from mice given an injection of vehicle (PBS) or 2 M NaCl.
(B) Differential enrichment of cell-type-specific genes in pS6 immunoprecipitates determined by Taqman. Data are expressed as the ratio of fold enrichment (IP/input) for salt-treated animals divided by the fold enrichment (IP/input) for controls and plotted on a log scale. Genes with a fold enrichment > 3.0 and p < 0.05 are labeled.
(C) Fold enrichment (IP/input) for a panel of activity-dependent genes following osmotic stimulation. Actin is shown for reference.
(D) RNA-seq analysis of differential enrichment of cell-type-specific genes in pS6 immunoprecipitates. Inset, magnification of region (blue) showing >4-fold enrichment. Genes labeled in (B) are highlighted.
(E) Colocalization between Avp, Oxt, and Crh with pS6 244 in salt-treated and control animals. Crh neurons were analyzed as two separate populations in the rostral and caudal PVN.
(F) pS6 intensity within individual Avp, Oxt, and Crh neurons from salt-treated and control animals.
(G) Colocalization between FosB, Cxcl1, and pS6 in salt-treated and control animals.
(H) Percentage of FosB-positive cells in the PVN and SON that are also pS6 positive.
(I) Percentage of pS6-positive cells in the PVN and SON that are also FosB positive.
All scale bars, 50 mm except (A) (200 mm). All error bars are mean ±SEM. See also Figure S3.