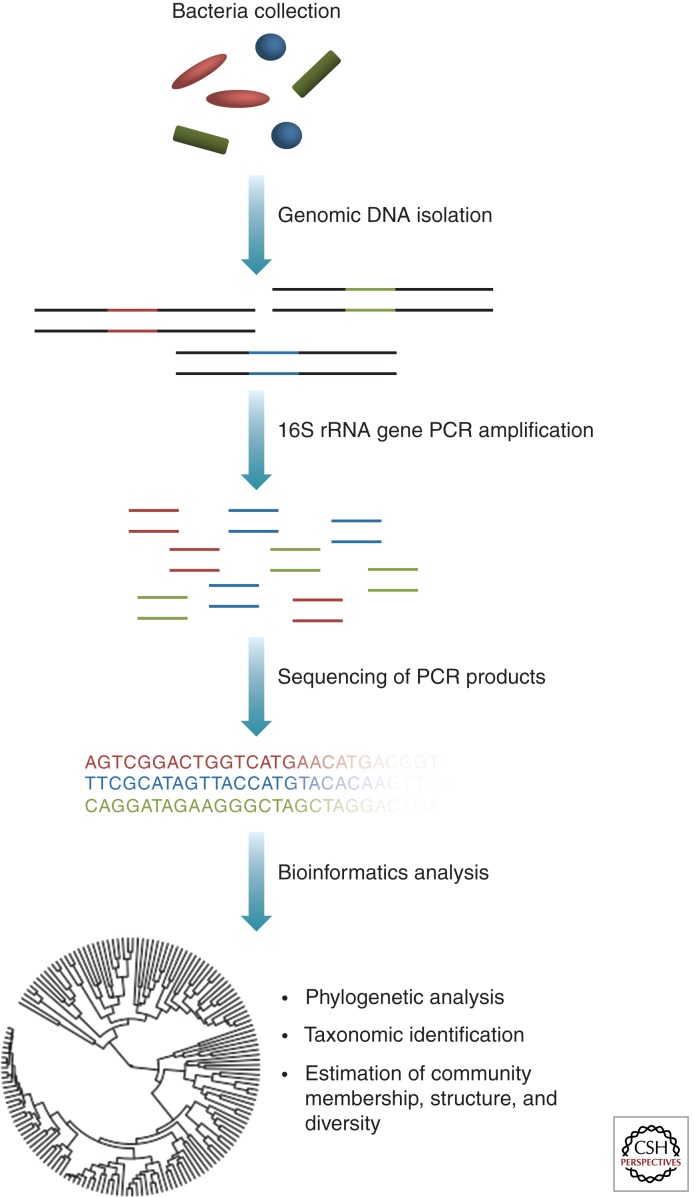
Figure 1.
The workflow of a bacterial 16S rRNA gene microbiome study. A heterogeneous mixture of genomic DNA is extracted from samples taken from the skin. Primers, containing barcodes that allow for multiplexing, are designed to the desired region of the 16S rRNA gene. 16S rRNA gene PCR products are amplified and sequenced. Low-quality sequences are removed, and various analyses are performed. These analyses can include assignment to taxonomy, analysis of shared phylogeny, and analysis of microbial community membership, structure, and diversity.