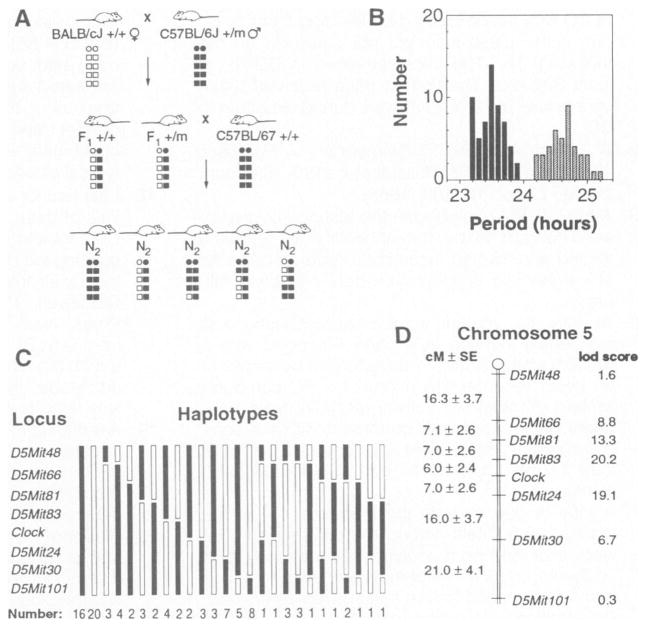
Fig. 4. Mapping of the Clock mutation.
A panel of 100 [(BALB × B6)F1 × B6]N2 mice was used to map Clock by genetic linkage and haplotype analysis. (A) The mating system used to produce the backcross panel. A schematic illustration of a pair of chromosomes is shown for each animal: circles represent centromeres, squares represent alleles at loci along the length of the chromosome, open symbols represent BALB alleles, and filled symbols represent B6 alleles. Clock heterozygous (+/m) B6 males were bred with wild-type (+/+) BALB females. The offspring were behaviorally tested and the Clock heterozygous F1 individuals were backcrossed to wild-type B6 to produce the N2 generation. One chromosome of each pair of autosomes in the N2 offspring is of B6 provenance (from the B6 parent), whereas the other (from the F1 parent) may be of either parental type or recombinant. Because the Clock mutation descends from a B6 animal, N2 offspring that are Clock/+ should carry B6 alleles at loci that flank the Clock locus, while +/+ offspring should carry BALB alleles at these loci. (B) Distribution of the period phenotype among the 100 animals. The steady-state period clearly distinguished animals between the two genotypes: 58 individuals were wild type and 42 individuals were heterozygous. See Table 1, cross 7, for analysis. (C) The assortment of haplotypes for chromosome 5 observed among the 100 N2 generation mice. Thirty-six animals had nonrecombinant haplotypes, 48 had singly recombinant haplotypes, and 16 exhibited doubly recombinant haplotypes. (D) Map of chromosome 5 showing the position of Clock relative to SSLP markers. Distances between loci are shown to the left in centimorgans ± standard error (cM ± SE), calculated with the program Map Manager (31). The lod scores to the right of each locus represent the logarithm of the odds for the pairwise test of linkage of the locus to Clock.