Significance
Duchenne muscular dystrophy (DMD) affects 1 in 3,500 live male births and is a fatal degenerative muscle disease with no known cure. The primary cause of DMD is muscle necrosis due to the loss of the dystrophin protein in the muscle membrane. However, the underlying cellular mechanisms that lead to cell death are not known. Using Caenorhabditis elegans as a model of muscular dystrophy, we show that whereas loss of dystrophin function is a primary cause of muscle degeneration, muscle cell death is greatly influenced by age-dependent, intrinsically variable cellular environments. We further show that reduction of insulin-like growth factor 1 signaling, which rejuvenates the cellular environments, helps to protect against muscle cell death caused by loss of dystrophin function.
Keywords: genetics, stress response, daf-2
Abstract
Duchenne muscular dystrophy, a fatal degenerative muscle disease, is caused by mutations in the dystrophin gene. Loss of dystrophin in the muscle cell membrane causes muscle fiber necrosis. Previously, loss-of-function mutations in dys-1, the Caenorhabditis elegans dystrophin ortholog, were shown to cause a contractile defect and mild fiber degeneration in striated body wall muscle. Here, we show that loss of dystrophin function in C. elegans results in a shorter lifespan and stochastic, age-dependent muscle-cell death. Reduction of dystrophin function also accelerated age-dependent protein aggregation in muscle cells, suggesting a defect in proteostasis. Both muscle cell death and protein aggregation showed wide variability among the muscle cells. These observations suggest that muscle cell death in dys-1 mutants is greatly influenced by cellular environments. Thus, the manipulation of the cellular environment may provide an opportunity to thwart the cell death initiated by the loss of dystrophin. We found that reduced insulin-like growth factor (IGF) signaling, which rejuvenates the cellular environment to protect cells from a variety of age-dependent pathologies, prevented muscle cell death in the dys-1 mutants in a daf-16–dependent manner. Our study suggests that manipulation of the IGF signaling pathways in muscle cells could be a potent intervention for muscular dystrophy.
Duchenne muscular dystrophy (DMD) is the most common form of muscular dystrophy, affecting 1 in 3,500 live male births. DMD is characterized by progressive muscle weakness and wasting with a consequential loss of mobility and fatality (1, 2). DMD is caused by mutations in the dystrophin gene. Dystrophin forms a complex at the muscle membrane with a number of proteins collectively termed the dystrophin-associated protein complex (DAPC). The DAPC is thought to provide mechanical stability to the muscle membrane through its interaction with the actin cytoskeleton and to function as a scaffold for signaling molecules. Loss of functional DAPCs at the muscle membrane leads to muscle necrosis, the primary cause of muscle degeneration. Although the cellular mechanisms underlying muscle necrosis are not clearly defined, mounting evidence indicates that cellular stress response and growth factor signaling play roles in the pathogenesis of DMD. For instance, muscles from DMD patients and from dystrophin-defective mdx mice have been shown to have aberrant activation of signaling molecules, such as AKT and mTOR (3, 4), and elevated antioxidant and heat shock protein (Hsp) levels (5–8). Myocytes isolated from mdx mice were shown to be more susceptible to oxidative injury (9).
The nematode Caenorhabditis elegans is a model organism that has profoundly contributed to our understanding of many cellular mechanisms relevant to human diseases (10–12). The C. elegans genome encodes a dystrophin ortholog, dys-1 (13), and mutations in the dys-1 gene lead to dysfunctional muscle contraction and mild muscle fiber degeneration (13, 14).
The forkhead box O (FoxO) family proteins are transcription factors that integrate growth factor signaling and cellular stress responses (15, 16). Their activities are largely regulated by nuclear-cytoplasmic shuttling that is mediated by posttranslational modifications, such as phosphorylation and acetylation; nuclear translocation is indicative of their activation (15, 16). Growth factors, such as insulin-like growth factor 1 (IGF1), activate the AKT kinase, which phosphorylates FoxO, thereby inhibiting its translocation into the nucleus; on the other hand, cellular stresses, such as oxidative stress, activate FoxO. C. elegans has a single FoxO ortholog, daf-16, and a single insulin/IGF receptor, daf-2 (17, 18). Reduced daf-2 activity results in elevated stress resistance and lifespan extension through robust daf-16 activation. Rejuvenation by reduced insulin/IGF1 activity also protects against age-dependent degenerative pathologies in C. elegans and mouse models of Alzheimer’s disease (10, 19).
In this study, we observed that C. elegans dystrophin mutants exhibit stochastic muscle cell death in an age-dependent manner. In an attempt to identify the underlying cellular mechanisms that lead to muscle cell death, we discovered that dys-1 mutation results in the disruption of proteostasis in muscle cells. Reduced IGF signaling can protect the muscle cells from death in a daf-16–dependent manner. These data indicate that the manipulation of the IGF signaling pathway may provide unique therapeutic strategies for muscular dystrophy.
Results
Premature Loss of Locomotory Function and Shortened Lifespan Are Observed in dys-1 Mutants.
Throughout this study, we examined two different dys-1 mutants alleles, dys-1(cx18) and dys-1(eg33), which were independently isolated by two different laboratories. The cx18 allele has a nonsense mutation at amino acid position 2721, and the eg33 allele has a nonsense mutation at position 3287 (Fig. S1). Although it has not been determined if these two alleles produce truncated proteins in vivo, it is likely that both of the resulting proteins have lost the scaffolding function at the muscle membrane because both alleles cause the mislocalization of dystrophin-associated proteins and the calcium activated BK (big K) channels at the muscle membrane (20, 21).
To examine whether the C. elegans dys-1 mutants exhibit a progressive loss of locomotory function, we measured the average speed of the wild-type and dys-1 mutant animals. Although the locomotory functions of both the wild-type and dys-1 animals declined progressively as they aged, this decline began earlier in the dys-1 mutants than in the wild-type animals (Fig. 1A). Because locomotory function is a good indicator of aging (22, 23), the rapid decline in the locomotory function of the dys-1 mutant prompted us to compare the lifespan of the wild-type and dys-1 animals. Indeed, the dys-1 animals have a shorter lifespan than the wild-type animals (Fig. 1B).
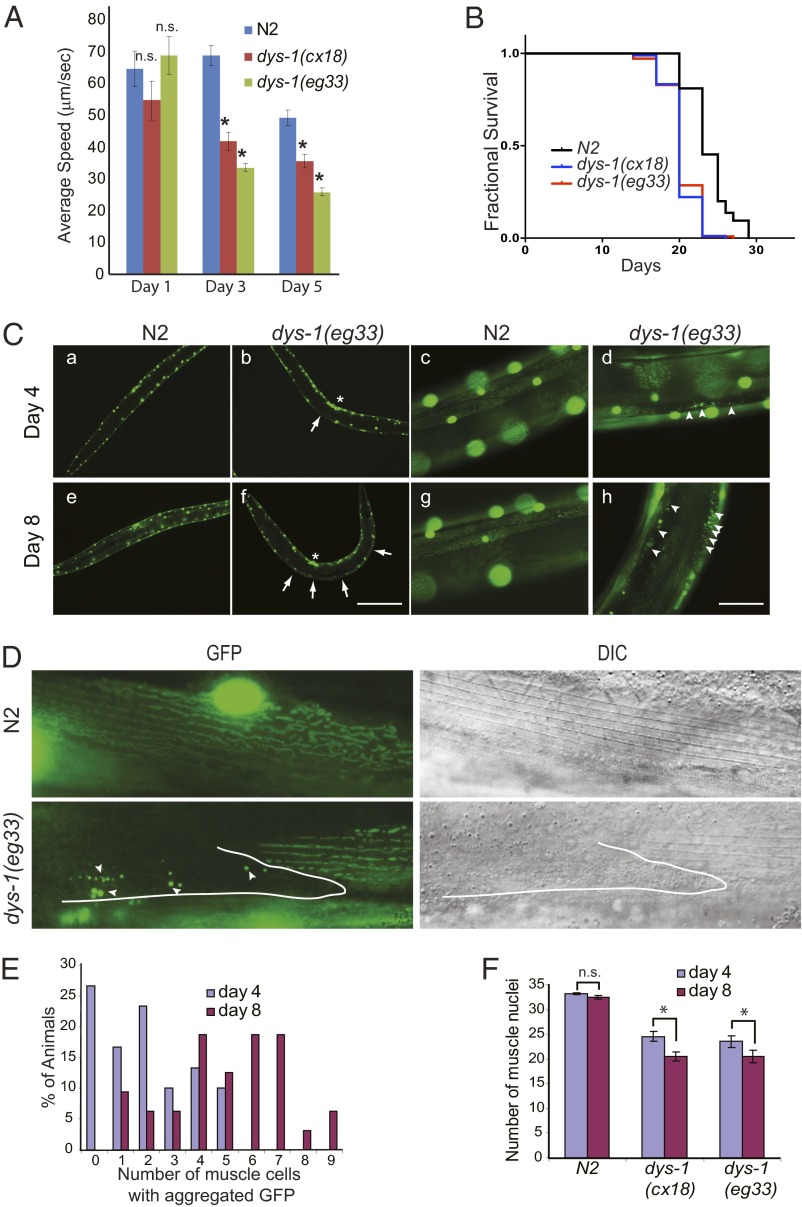
Fig. 1.
dys-1 mutant animals exhibit a premature decline of locomotory function and shortened lifespan. (A) Average speed of the wild-type (N2) and dys-1(eg33) and dys-1(cx18) mutant animals on food. The L4 stage was set as day 0. Error bars represent SEM. *P < 0.005; n.s., not significant compared with N2; two-tailed Student t test; n > 30. (B) dys-1 mutants have a shorter lifespan than the wild-type (N2) animals. The L4 stage was set as day 0. Animals were grown on OP50 at 20 °C. Survival data were analyzed using the Kaplan–Meier method. For statistical data and additional independent trials, see Table S1. (C) Muscle cell death was observed in the dys-1 mutant, but not in the wild-type animals. Wild-type and dys-1 animals expressing GFP in the muscle nuclei and mitochondria (ccIs4251[Pmyo-3GFP-NLS, Pmyo-3 GFP-mit]) were grown synchronously at 20 °C and were observed at the indicated time points. The arrows indicate muscle cells that have lost the distinct circular nuclear GFP signals. The asterisk indicates the vulva. Arrowheads indicate the areas that show aggregated GFP, which is indicative of cell death. Under low magnification, GFP signals from the mitochondrial networks could not be observed. (Scale bars: 20 μm in a, b, e, and f, and 5 μm in c, d, g, and h.) (D) Muscle cells with aggregated GFP had lost the distinctive dense body pattern. Upper and Lower Left show the GFP fluorescence images of muscle cells, and Upper and Lower Right show DIC images of the same muscle cells. Arrowheads indicate aggregated GFP, and the white outline demarcates the boundary of the muscle cell. (E) The number of dead muscle cells in the dys-1 mutants increased with age. The number of dead muscle cells in the dys-1 animals was scored by counting the muscle cells with aggregated GFP at days 4 and 8. P < 0.0001, χ2; n = 30 for day 4 and n = 32 for day 8. (F) The average number of observable dys-1 muscle cell nuclei declined as the animals aged due to muscle cell death. The number of muscle nuclei located in a portion of two dorsal quadrants, as shown in Fig. S3 was counted; typically, 30 muscle nuclei were observed in the wild-type animals. Error bars represent SEM. *P < 0.0001, two-tailed Student t test; n > 20.
Muscle Cell Death in dys-1 Mutants Is Age Dependent.
One possible explanation for the early decline in the locomotory function of the dys-1 mutants is the accumulation of muscle cell death or damage as they age. To test this possibility, we compared the structural/morphological integrity of the wild-type and dys-1 muscle cells as a function of age. We crossed the dys-1 animals with a transgenic strain expressing GFP that is targeted to the nucleus and mitochondria in a muscle-specific manner. Nucleus-targeted GFP has been used as an indicator for cell survival and death (24). We also included the mitochondria-targeted GFP because mitochondria play crucial roles in locomotory function and the survival of muscle cells. Importantly, abnormal mitochondrial function is a common feature of various types of muscular dystrophy (25–28).
Synchronously growing animals were observed periodically (every 2 or 3 d) over their lifetime under a fluorescent microscope. At day 4 of adulthood, wild-type animals showed strong GFP signals throughout the nuclei of whole body wall muscle cells, whereas ∼70% of the dys-1 mutant animals showed a loss of regularly spaced nuclear GFP signals in some muscle cells when examined under low magnification (Fig. 1C). When we examined the animals under a higher magnification, the muscle nuclei of the wild-type animals showed strong GFP signals, and the mitochondria were arranged in mesh-like structures distributed underneath the myofilament lattice (Fig. 1D). In contrast, the dys-1 mutant muscle cells showed patches of aggregated GFP without discernible nuclei or mitochondria (Fig. 1 C and D), which resembled the features of necrotic cells (24). To further confirm that the muscle cells containing aggregated GFP were dead or dying cells, we examined them under a differential interference contrast (DIC) microscope. Live muscle cells exhibited typical dense body patterns (Z lines/costameres), but the cells with aggregated GFP no longer showed such a pattern (Fig. 1D). Such abnormal GFP signals in the dys-1 muscle cells did not result from changes in the nuclear GFP expression levels because when we stained the nuclei of the dys-1 mutants with DAPI, we found that the abnormal GFP signals coincided with nuclear shrinkage and fragmentation (Fig. S2). Together, we concluded that the absence of nuclear GFP or aggregated GFP in the muscle cells represented dead cells, and the pattern of nuclear GFP serves as a good indicator of muscle cell health. On day 8 of adulthood, the wild-type animals still showed strong GFP signals in the nuclei and mitochondria (Fig. 1C). The most striking features of the dys-1 animals were an increase in the proportion of the animals exhibiting muscle cell death and an increase in the number of dead muscle cells in the affected animals (Fig. 1E). As a consequence, the average number of observable muscle nuclei in the dys-1 animals decreased considerably from day 4 to day 8 (Fig. 1F). Importantly, muscle cells containing aggregated GFP were not observed in the wild-type animals even at day 18 (Fig. S4), which is consistent with an earlier report that showed that nuclei rarely break down, even at very old age (22). These data suggest that the early decline of the locomotory function in the dys-1 animals was due, in large part, to muscle cell death, even though we cannot completely rule out the involvement of accelerated aging. It is noteworthy that even though each body wall muscle cell was defective in dystrophin, the affected muscle cells were random in their position and lineage (Fig. 1C). In the dys-1 mutant, a dead muscle cell was often surrounded by normal, healthy muscle cells (Fig. 1D). Together, these results strongly suggest that the observed cell death is not mediated by the loss of dystrophin alone; rather, it is greatly influenced by age-dependent, intrinsically variable cellular environments.
Dystrophin Knock-Down Leads to the Disruption of Protein Homeostasis.
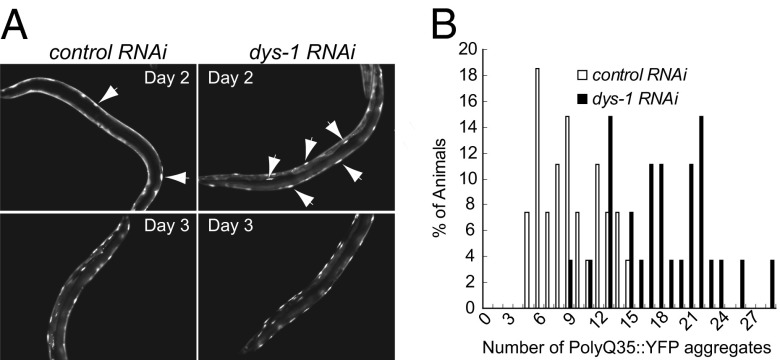
Muscles are under constant mechanical, thermal, and oxidative stresses and are highly dependent on protein quality control systems to maintain muscular function (29, 30). Garcia et al. reported that hyperexcitation by GABAergic and cholinergic signaling in postsynaptic muscles causes an imbalance in protein homeostasis that is, at least in part, due to calcium overload (31). The dystrophin-deficient body wall muscles of C. elegans are hyperexcitable (13, 21), and stress proteins and calcium levels are elevated in DMD and mdx muscles (7). To examine the status of protein homeostasis in the muscle cells, we took advantage of the polyglutamine (polyQ) aggregation model in C. elegans developed by Morley et al. (32). Protein homeostasis in aging muscle cells was shown to be progressively compromised (32, 33). Furthermore, hyperexcitation of muscle cells accelerates and exacerbates polyQ aggregation, leading to polyQ aggregation at younger ages (31, 32). We reasoned that if the dys-1 mutants exhibited disrupted protein homeostasis, we should be able to observe an earlier onset of protein aggregation in their muscle cells. We fed animals expressing polyQ35::YFP (AM140) in the body wall muscles with bacteria harboring either a control (empty vector) or dys-1 RNAi-feeding vector and compared the severity and onset of polyQ35 aggregation. The effectiveness of dys-1 knock-down was demonstrated by the appearance of the head-bending phenotype (hypercontracted anterior body part), which is a unique locomotory phenotype of the dys-1 mutant (13). We found that polyQ aggregation appeared earlier in the dys-1 RNAi-treated animals than in the control RNAi-treated animals, suggesting that the loss of dys-1 function disrupted protein homeostasis in muscle cells (Fig. 2 A and B). We also observed a wide variation in the extent of YFP aggregation among the individual animals and among the muscle cells in individual animals (Fig. 2B). To eliminate the possibility of variable RNAi effects, we repeated the experiment with dys-1 mutants crossed to AM140 animals and obtained the same result (Fig. S5). This observation suggests that, similar to muscle cell death in the dys-1 mutant animals, protein homeostasis is highly influenced by stochastic cellular environments.
Fig. 2.
Knock-down of dys-1 accelerates age-dependent protein aggregation in muscle cells. (A) Protein aggregation in the muscle cells was examined using transgenic animals expressing polyQ35::YFP in the body wall muscles (AM140), which shows age-dependent aggregation (32). The AM140 strain was fed with bacteria harboring either a control (empty vector) or dys-1 RNAi-feeding vector. The L4 stage was set for day 0. The arrows indicate the polyQ35::YFP aggregates. (Magnification: 10×.) (B) PolyQ35::YFP aggregates were quantified in the control (white bars) and dys-1 (black bars) RNAi-treated AM140 animals at day 3 of adulthood. P < 0.0001, χ2; n = 27. Three independent experiments produced similar results.
The stochastic nature of protein aggregation in muscle cells led us to investigate whether cellular stress levels in the muscle cells vary as well. We examined the expression level of a gst-4 transcriptional reporter, which is induced by oxidative stresses and the collapse of proteostasis (34, 35). In both the wild type and dys-1 animals, gst-4 expression was variable among the individual muscle cells of a given animal (Fig. S6A). This also shows that even though the muscle cells in a given animal are genetically identical, they exhibit variable physiologies. It was difficult to compare the gst-4 reporter expression level between the different strains due to the variability in the muscle cells and the high level of expression in the gut and other tissues. However, we observed higher overall gst-4 reporter expression in the dys-1(eg33) mutant, but not in the dys-1(cx18) mutant, compared with the wild-type animals at the young adult stage; however, this difference was not observed in the older animals (Fig. S6 B and C). Treating the animals with dys-1 RNAi resulted in an overall increase in gst-4::GFP expression at least in the gut, suggesting that the loss of dys-1 function may have impacts on multiple tissues (Fig. S7). The lack of impact of the cx18 allele on gst-4::GFP reporter expression may be due to an imprecise measurement or an effect of the genetic background. When we examined another stress response transcription factor, DAF-16, we found that DAF-16 translocated into the nuclei of the muscle cells (not other tissues) in the dys-1(eg33) mutant briefly for about 6 h (when examined under a dissecting fluorescent microscope); however, this was not observed in the dys-1(cx18) mutant (Fig. S8A). Treating the DAF-16::GFP animals with the dys-1 RNAi did not result in the nuclear localization of DAF-16 at the young adult stage nor did it abolish the transient nuclear localization of DAF-16 in the eg33 allele (Fig. S8B); therefore, it appears that the eg33 allele has a genetic background that is more sensitive to stress. Despite this difference between the cx18 and eg33 alleles at the young adult stage, both alleles show similar levels of muscle cell death (Fig. 1). Thus, the brief DAF-16 activation and the higher gst-4 expression in the dys-1(eg33) mutant at the young adult stage did not have a significant impact on the survival of the muscle cells.
Reduced IGF Signaling Protects the dys-1 Mutant from Muscle Cell Death.
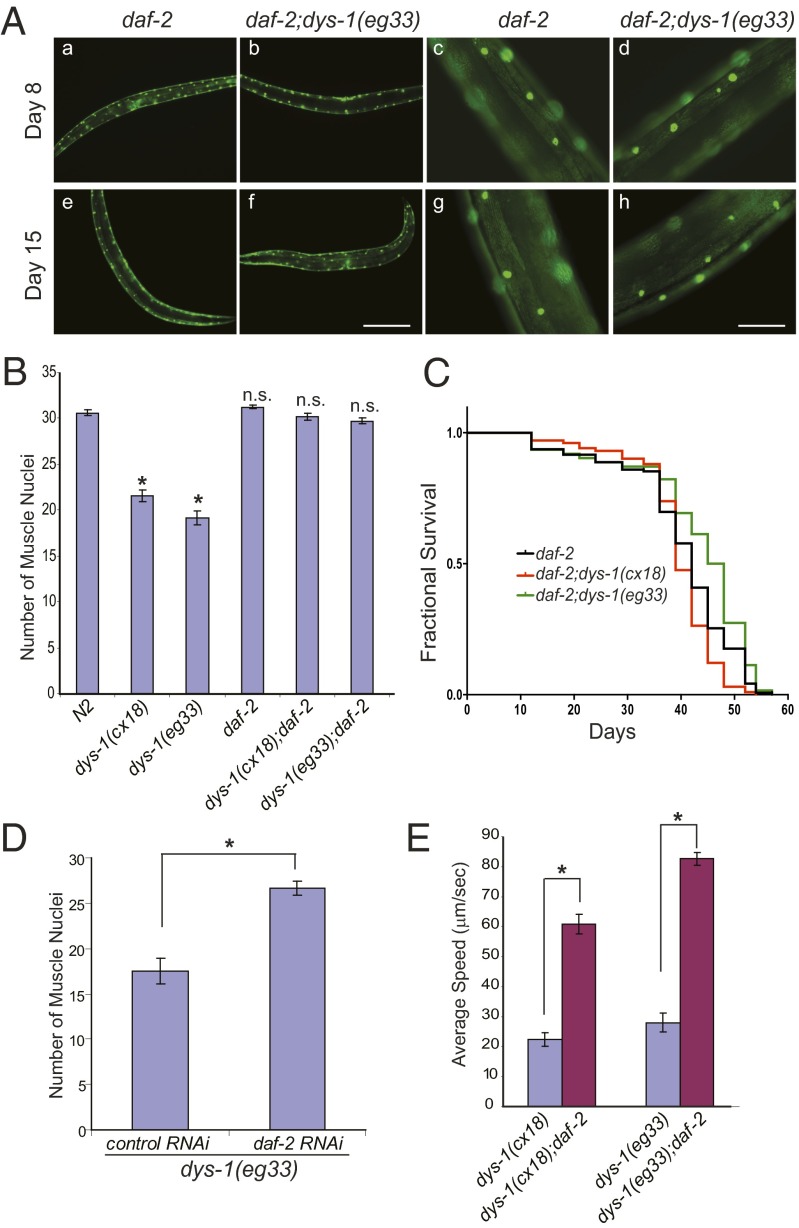
Our data show that the loss of dystrophin function does not directly cause muscle cell death. Rather, it causes proteostasis stress in the muscle cells. Muscle cell death or survival in the dys-1 mutants is determined by the cellular environment, which varies among the individual muscle cells and is age-dependent. This conclusion led us to reason that a sustained, rejuvenating cellular environment may provide protection from muscle cell death. It is well known that reductions in IGF signaling can enhance stress resistance, delay inherent protein aggregation, and extend lifespan (11). These beneficial effects on age-dependent degenerative phenotypes are largely dependent on daf-16 (36). After crossing the dys-1 mutant with the daf-2(e1370) mutant, which has a reduction-of-function mutation in the gene encoding an IGF receptor homolog, we assessed the health of the muscle cells and measured the animals’ lifespans. Indeed, reduced IGF signaling protected the dys-1 mutant from muscle cell death, and we were not able to observe the aggregated GFP in the muscle cells, even at day 15 of adulthood (Fig. 3 A and B). Additionally, the lifespan of the daf-2;dys-1 double mutants was extended to a level similar to the daf-2 mutant alone (Fig. 3C). This protective effect was also recapitulated by the reduction of DAF-2 using RNAi (Fig. 3D). To examine whether the protective effect of reduced DAF-2 signaling on muscle cell death and aging was translated into muscle function, we compared the speed of the dys-1 and dys-1;daf-2 mutants and found that the dys-1;daf-2 mutants had much more robust mobility (Fig. 3E and Movies S1 and S2).
Fig. 3.
Reduced DAF-2 function prevents muscle cell death in the dys-1 mutants. (A) A reduction-of-function mutation, daf-2(e1370), improved the muscle pathology of the dys-1 mutant. Aggregated GFP was not found in the muscle cells of the dys-1(eg33);daf-2(e1370) and daf-2(e1370) animals on days 8 and 15 of adulthood. Muscle cells were visualized with ccIs4251 as in Fig. 1. The animals were grown synchronously at 20 °C. (Scale bars: 20 μm in a, b, e, and f, and 5 μm in c, d, g, and h.) (B) The average number of muscle nuclei was quantified, as in Fig. 1F (n = 20). Error bars represent SEM. *P < 0.0001 compared with N2; n.s., not significant compared with N2; two-tailed Student t test. (C) daf-2(e1370) extended the lifespan of the dys-1 mutant animals. Survival of the daf-2(e1370), daf-2(e1370);dys-1(eg33), and daf-2(e1370);dys-1(cx18) animals was analyzed using the Kaplan–Meier method. For statistical data and additional independent trials, see Table S1. (D) Reduction of DAF-2 by RNAi feeding protected the dys-1(eg33) mutant from the loss of muscle nuclei. The dys-1(eg33) animals were treated with either a control (empty vector, n = 20) or daf-2– (n = 15) feeding RNAi, and the muscle nuclei were counted on day 4 of adulthood. Error bars represent SEM. *P < 0.0001, two-tailed Student t test. (E) A reduction-of-function mutation, daf-2(e1370), improved the mobility of the dys-1 mutants. Day-7 adulthood animals were transferred to freshly seeded plates, and their speeds were measured immediately as in Fig. 1A. Error bars represent SEM. *P < 0.0001, two-tailed Student t test; n > 20. For video, see Movies S1 and S2.
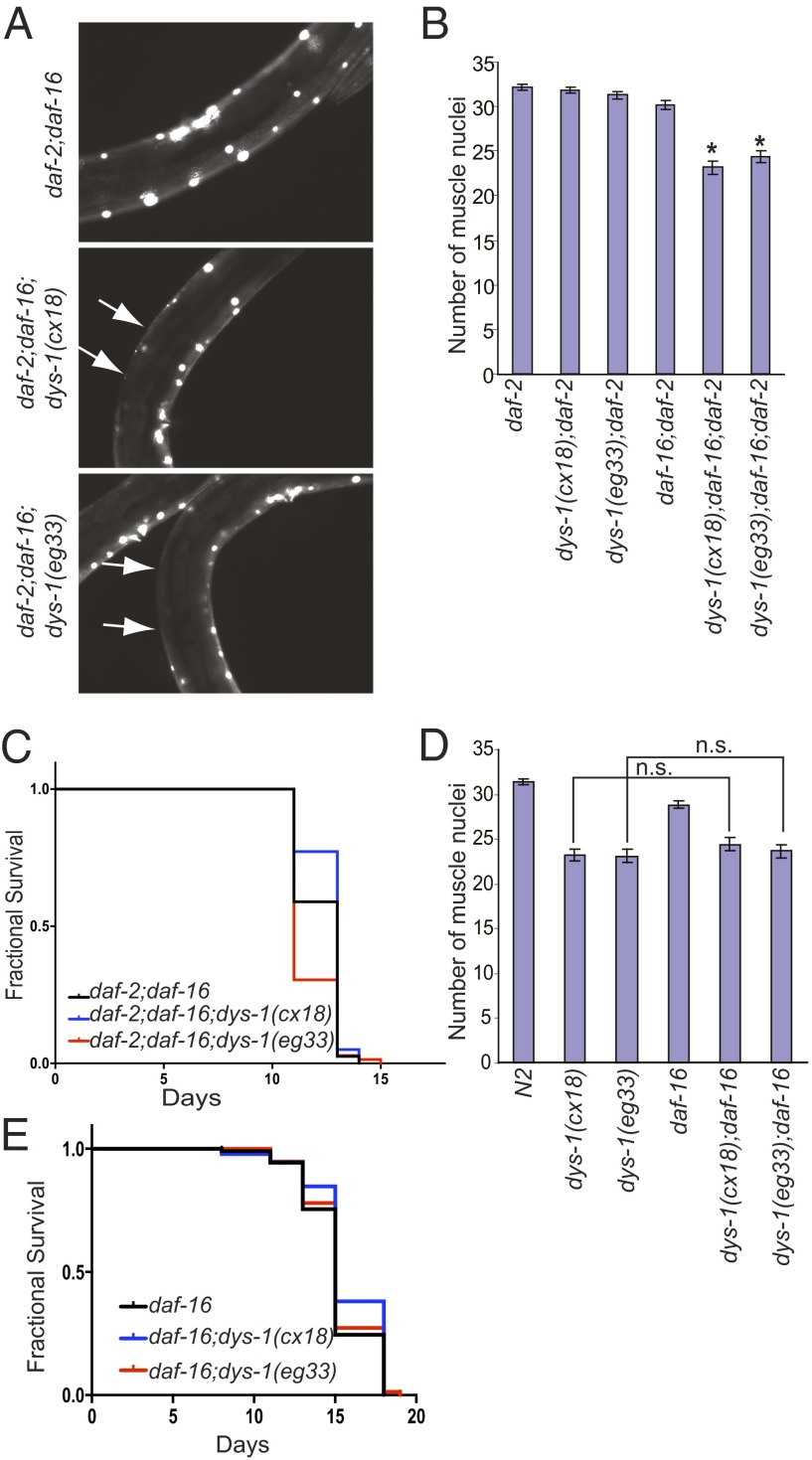
Next, we asked if the protective effects of reduced IGF signaling were dependent on DAF-16. We generated triple mutants daf-2(e1370);daf-16(mu86);dys-1. In these triple mutants, the aggregated GFP reappeared, and the number of muscle nuclei was reduced (Fig. 4 A and B). Similar results were obtained when we used RNAi feeding to reduce the expression of daf-16 (Fig. S9). The lifespan of the triple mutants (daf-2;daf-16;dys-1) was similar to that of the daf-2;daf-16 double mutant (Fig. 4C). This result indicated that the beneficial effect of reduced IGF signaling on the muscle cells of the dys-1 animals was largely dependent on DAF-16. We then asked if DAF-16 played a major role in the maintenance of muscle cell viability in the dys-1 mutant under normal IGF signaling conditions. When we compared the extent of muscle cell death in the dys-1 mutant with the dys-1;daf-16 double mutant, no difference was observed (Fig. 4E). Additionally, the lifespan of the daf-16 single mutant was not different from the daf-16;dys-1 double mutant (Fig. 4E). It appears that, in a normal IGF signaling environment, where DAF-16 is inhibited, the survival of the muscle cells does not depend on DAF-16. However, under reduced IGF signaling conditions, DAF-16 is required to activate survival mechanisms in the muscle cells of the dys-1 mutants.
Fig. 4.
DAF-16 is necessary for the protective effect of reduced IGF signaling on muscle cells but is not required for the survival of the muscle cells under normal IGF signaling conditions. (A) daf-16 is required for the protective effect of the daf-2(e1370) mutation. Muscle cell death reappeared in the triple mutant, daf-2(e1370);daf-16(mu86);dys-1. The arrows indicate the areas with lost muscle nuclei; 20× objective was used for image acquisition. (B) Quantification of the average muscle cell nuclei. Error bars represent SEM. *P < 0.0001 compared with daf-2, two-tailed Student t test; n = 20. (C) The lifespan of the triple mutant daf-2(e1370);daf-16(mu86);dys-1 was not different from that of the double mutant, daf-2(e1370);daf-16(mu86). Survival data were analyzed using the Kaplan–Meier method. For statistical data and additional independent trials, see Table S1. (D) Loss of DAF-16 did not exacerbate the muscle cell death in the dys-1 mutant under normal DAF-2 function. The average muscle cell nuclei of the double mutant, dys-1;daf-16, were compared with the single mutant, dys-1. Error bars represent SEM. n.s., not significant; two-tailed Student t test; n = 20. (E) The lifespan of the double mutant daf-16(mu86);dys-1 was not different from that of daf-16(mu86). Survival data were analyzed using the Kaplan–Meier method. For statistical data and additional independent trials, see Table S1.
Discussion
Loss of dystrophin has been shown to compromise the integrity of muscle cells and cause calcium overload in the muscles of humans and other model organisms, including C. elegans (1, 7, 37). In this study, we showed that C. elegans dystrophin mutants also exhibited muscle cell death in an age-dependent manner. Importantly, the affected muscle cells were random in their positions and lineage. This implies that although the primary cause of muscle cell death is the loss of dystrophin function, muscle cell death in dys-1 mutant animals is highly influenced by the downstream cellular responses that are highly variable for individual cells.
Proteostasis is crucial for maintaining muscle integrity and is regulated by the cellular stress response (29). We used the polyQ35 aggregation model to probe the status of protein homeostasis in the muscles of the mutant animals. As the animals become older, the capacity to maintain protein-folding homeostasis diminishes, resulting in an increase in polyQ35 aggregation (32). We found that dystrophin knock-down accelerated polyQ35 aggregation in the muscle cells. This indicates that the loss of dystrophin compromises the capacity of these cells to maintain protein-folding homeostasis. Compromised protein-folding capacity can lead to the misfolding and dysfunction of structurally unstable proteins that are tolerated under normal conditions (38). In this context, it is worth noting that the elevation of Hsp72 expression, either by transgenics or by treatment with the pharmacological inducer of Hsp72 (BGP-15, a hydroxylamine derivative), ameliorates the dystrophic phenotypes of mdx mice (39). Hsp72 is induced by stress and is critical for maintaining proteostasis by enhancing the cellular folding capacity (40). Genetic background is known to influence the pathogenesis of muscular dystrophy (41, 42), although the genetic modifiers that underlie the phenotypic variability of muscular dystrophy are not known. It is intriguing to speculate that certain polymorphisms may destabilize protein structures, and their contributions to pathogenesis only manifest when the primary causal gene of muscular dystrophy is mutated, resulting in disrupted proteostasis.
Recent studies have demonstrated that muscles play important roles in regulating whole body homeostasis. FoxO overexpression in muscles was shown to preserve protein homeostasis during aging in multiple tissues and to extend the lifespan of the organism, and the overexpression of the aggregation-prone human Huntington disease protein in muscles was shown to shorten lifespan in Drosophila (43). Perturbation of proteostasis in muscle cells was reported to induce HSP90 expression in multiple tissues (44). Consistent with the global impact of muscle physiology, we observed that knock-down of dystrophin led to the induction of the gst-4::GFP reporter in the intestine, and the dystrophin mutation shortens the lifespan of C. elegans.
We found that reducing IGF signaling, which is known to rejuvenate cells through a variety of mechanisms that are directly or indirectly dependent on DAF-16, prevented muscle cell death in the dys-1 mutant. Our data are consistent with the role of DAF-16 as a mediator of the stress-resistance response that protects cells from damage and extends the lifespan of the organisms (16). In mammals, IGF1 is known to stimulate the proliferation of muscle stem cells, known as satellite cells, and to induce hypertrophy in skeletal muscles (45, 46). Therefore, IGF1 is considered a potential therapeutic agent for DMD. However, it is not clear whether IGF1 is beneficial in the treatment of muscular dystrophy. For instance, it was noted that elevated IGF signaling, which induces muscle hypertrophy, does not provide significant protection from muscle necrosis (46). Our data indicated that reducing IGF1 signaling enhanced the survival of the dystrophin-deficient muscle cells in C. elegans. Consistent with our finding, chronic activation of AMPK (AMP-activated protein kinase), which is a FoxO activator, was recently shown to increase sarcolemmal integrity, under basal conditions and during damaging eccentric contraction in mdx mice (47). Inhibition of TORC1, which is inhibited by FoxO under stress conditions (48), by rapamycin treatment was also shown to reduce muscle necrosis and fibrosis in the mdx mice (49).
Effective DMD treatments must prevent or delay muscle necrosis and enhance muscle regeneration until dystrophin replacement therapy becomes a reality (50). Continued necrosis will eventually exhaust the stem cell populations and lead to fatality (51). Given that both necrosis and regeneration are important aspects to consider for therapy, it will be important to dissect the IGF signaling pathway and to target the downstream signaling molecules that maximize the proper balance between muscle protection and regeneration.
Materials and Methods
Strains.
The following strains were obtained from the Caenorhabditis Genetic Center: N2, TJ356 (zIs356 [Pdaf-16 daf-16-gfp, rol-6(d)]), CB1370 (daf-2(e1370)), CF1038 (daf-16(mu86)), CB5600 (ccIs4251[Pmyo-3NLS-gfp-lacZ; Pmyo-3Mt-gfp]; him-8(e1489)), AM140 (rmIs132[Punc-54Q35-YFP]), dys-1(cx18), and CL2166 (dvIs19[pAF15(gst-4::GFP::NLS)]). N2 worms were crossed with CB5600 to generate transgenic animals expressing nuclear and mitochondrial GFP in the body wall muscles, resulting in the HKK90 strain. him-8(e1489) was eliminated during the crossing. HKK90 strain males were crossed with dys-1 mutants to generate dys-1 mutants expressing the GFP transgene in their body wall muscles. The dys-1 mutants were crossed with TJ356, resulting in the dys-1 mutant animals with the DAF-16::GFP transgene.
Lifespan Assay.
Eggs were collected by bleaching with sodium hypocholrite and were allowed to hatch in M9 buffer for 16 h. Synchronized L1-stage animals were transferred to seeded nematode growth media (NGM) plates. Animals 16–24 h post-L4 stage were transferred to seeded NGM plates containing 100 μg/mL 5-fluorodeoxy uridine (FUDR, Sigma), and live and dead animals were counted every 2–3 d. The assays were performed at 20 °C with the Escherichia coli strain (OP50) as a food source. Survival data were analyzed using the Kaplan–Meier method and Prism (version 6.0) software.
Measurement of the Average Speed.
The speed of the animals was measured on food. Movie frames were acquired using a dissecting microscope equipped with a Go-3 digital camera (QImaging) for 2 min with a 500-msec interval and 20-msec exposures. We measured the average speed of the animals using Track Objects from ImagePro Plus (Media Cybernetics).
Image Acquisition.
Animals were anesthetized with 25 mM sodium azide, mounted on agar pads, and imaged with a Zeiss AxioObserver microscope equipped with a fluorescent light source. MetaMorph 7.6 (Molecular Dynamics) was used to process the images.
PolyQ Aggregation Assay.
The polyQ aggregation assay was performed as previously described (31). The AM140 strain was grown on either control (empty vector) or dys-1–feeding RNAi (Ahringer RNAi Library, Source BioScience) plates. Animals were synchronized by allowing the adults to lay eggs for 1 h and then removing them from the plates. At the given time points, the animals were mounted on agar pads containing 1 mM levamisole, and the YFP::GFP aggregates were counted.
Supplementary Material
Acknowledgments
We thank Dr. Garsin for sharing a daf-2 RNAi clone, and J. Pierce-Shimomura and L. Abraham for critically reading the manuscript. Some strains were provided by the CGC, which is funded by National Institutes of Health Office of Research Infrastructure Programs (P40OD01440). This work was supported by the NIH Grant R21NS077018 (to H.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1308866110/-/DCSupplemental.
References
- 1.Goldstein JA, McNally EM. Mechanisms of muscle weakness in muscular dystrophy. J Gen Physiol. 2010;136(1):29–34. doi: 10.1085/jgp.201010436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davies KE, Nowak KJ. Molecular mechanisms of muscular dystrophies: Old and new players. Nat Rev Mol Cell Biol. 2006;7(10):762–773. doi: 10.1038/nrm2024. [DOI] [PubMed] [Google Scholar]
- 3.Peter AK, Crosbie RH. Hypertrophic response of Duchenne and limb-girdle muscular dystrophies is associated with activation of Akt pathway. Exp Cell Res. 2006;312(13):2580–2591. doi: 10.1016/j.yexcr.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 4.Feron M, et al. PTEN contributes to profound PI3K/Akt signaling pathway deregulation in dystrophin-deficient dog muscle. Am J Pathol. 2009;174(4):1459–1470. doi: 10.2353/ajpath.2009.080460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doran P, Martin G, Dowling P, Jockusch H, Ohlendieck K. Proteome analysis of the dystrophin-deficient MDX diaphragm reveals a drastic increase in the heat shock protein cvHSP. Proteomics. 2006;6(16):4610–4621. doi: 10.1002/pmic.200600082. [DOI] [PubMed] [Google Scholar]
- 6.Ge Y, Molloy MP, Chamberlain JS, Andrews PC. Proteomic analysis of mdx skeletal muscle: Great reduction of adenylate kinase 1 expression and enzymatic activity. Proteomics. 2003;3(10):1895–1903. doi: 10.1002/pmic.200300561. [DOI] [PubMed] [Google Scholar]
- 7.Tidball JG, Wehling-Henricks M. The role of free radicals in the pathophysiology of muscular dystrophy. J Appl Physiol (1985) 2007;102(4):1677–1686. doi: 10.1152/japplphysiol.01145.2006. [DOI] [PubMed] [Google Scholar]
- 8.Disatnik MH, et al. Evidence of oxidative stress in mdx mouse muscle: Studies of the pre-necrotic state. J Neurol Sci. 1998;161(1):77–84. doi: 10.1016/s0022-510x(98)00258-5. [DOI] [PubMed] [Google Scholar]
- 9.Rando TA, Disatnik MH, Yu Y, Franco A. Muscle cells from mdx mice have an increased susceptibility to oxidative stress. Neuromuscul Disord. 1998;8(1):14–21. doi: 10.1016/s0960-8966(97)00124-7. [DOI] [PubMed] [Google Scholar]
- 10.Cohen E, Bieschke J, Perciavalle RM, Kelly JW, Dillin A. Opposing activities protect against age-onset proteotoxicity. Science. 2006;313(5793):1604–1610. doi: 10.1126/science.1124646. [DOI] [PubMed] [Google Scholar]
- 11.Cohen E, Dillin A. The insulin paradox: Aging, proteotoxicity and neurodegeneration. Nat Rev Neurosci. 2008;9(10):759–767. doi: 10.1038/nrn2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Markaki M, Tavernarakis N. Modeling human diseases in Caenorhabditis elegans. Biotechnol J. 2010;5(12):1261–1276. doi: 10.1002/biot.201000183. [DOI] [PubMed] [Google Scholar]
- 13.Bessou C, Giugia JB, Franks CJ, Holden-Dye L, Ségalat L. Mutations in the Caenorhabditis elegans dystrophin-like gene dys-1 lead to hyperactivity and suggest a link with cholinergic transmission. Neurogenetics. 1998;2(1):61–72. doi: 10.1007/s100480050053. [DOI] [PubMed] [Google Scholar]
- 14.Giugia J, Gieseler K, Arpagaus M, Ségalat L. Mutations in the dystrophin-like dys-1 gene of Caenorhabditis elegans result in reduced acetylcholinesterase activity. FEBS Lett. 1999;463(3):270–272. doi: 10.1016/s0014-5793(99)01651-8. [DOI] [PubMed] [Google Scholar]
- 15.Calnan DR, Brunet A. The FoxO code. Oncogene. 2008;27(16):2276–2288. doi: 10.1038/onc.2008.21. [DOI] [PubMed] [Google Scholar]
- 16.van der Horst A, Burgering BM. Stressing the role of FoxO proteins in lifespan and disease. Nat Rev Mol Cell Biol. 2007;8(6):440–450. doi: 10.1038/nrm2190. [DOI] [PubMed] [Google Scholar]
- 17.Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278(5341):1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- 18.Ogg S, et al. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389(6654):994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 19.Cohen E, et al. Reduced IGF-1 signaling delays age-associated proteotoxicity in mice. Cell. 2009;139(6):1157–1169. doi: 10.1016/j.cell.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen B, et al. α-Catulin CTN-1 is required for BK channel subcellular localization in C. elegans body-wall muscle cells. EMBO J. 2010;29(18):3184–3195. doi: 10.1038/emboj.2010.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim H, et al. The dystrophin complex controls bk channel localization and muscle activity in Caenorhabditis elegans. PLoS Genet. 2009;5(12):e1000780. doi: 10.1371/journal.pgen.1000780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herndon LA, et al. Stochastic and genetic factors influence tissue-specific decline in ageing C. elegans. Nature. 2002;419(6909):808–814. doi: 10.1038/nature01135. [DOI] [PubMed] [Google Scholar]
- 23.Collins JJ, Huang C, Hughes S, Kornfeld K. The measurement and analysis of age-related changes in Caenorhabditis elegans. WormBook. 2008;24:1–21. doi: 10.1895/wormbook.1.137.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scott BA, Avidan MS, Crowder CM. Regulation of hypoxic death in C. elegans by the insulin/IGF receptor homolog DAF-2. Science. 2002;296(5577):2388–2391. doi: 10.1126/science.1072302. [DOI] [PubMed] [Google Scholar]
- 25.Kuznetsov AV, et al. Impaired mitochondrial oxidative phosphorylation in skeletal muscle of the dystrophin-deficient mdx mouse. Mol Cell Biochem. 1998;183(1-2):87–96. doi: 10.1023/a:1006868130002. [DOI] [PubMed] [Google Scholar]
- 26.Bernardi P, Bonaldo P. Dysfunction of mitochondria and sarcoplasmic reticulum in the pathogenesis of collagen VI muscular dystrophies. Ann N Y Acad Sci. 2008;1147:303–311. doi: 10.1196/annals.1427.009. [DOI] [PubMed] [Google Scholar]
- 27.Durieux J, Dillin A. Mitochondria and aging: Dilution is the solution. Cell Metab. 2007;6(6):427–429. doi: 10.1016/j.cmet.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 28.Millay DP, et al. Genetic and pharmacologic inhibition of mitochondrial-dependent necrosis attenuates muscular dystrophy. Nat Med. 2008;14(4):442–447. doi: 10.1038/nm1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arndt V, et al. Chaperone-assisted selective autophagy is essential for muscle maintenance. Curr Biol. 2010;20(2):143–148. doi: 10.1016/j.cub.2009.11.022. [DOI] [PubMed] [Google Scholar]
- 30.Frank D, Kuhn C, Katus HA, Frey N. Role of the sarcomeric Z-disc in the pathogenesis of cardiomyopathy. Future Cardiol. 2007;3(6):611–622. doi: 10.2217/14796678.3.6.611. [DOI] [PubMed] [Google Scholar]
- 31.Garcia SM, Casanueva MO, Silva MC, Amaral MD, Morimoto RI. Neuronal signaling modulates protein homeostasis in Caenorhabditis elegans post-synaptic muscle cells. Genes Dev. 2007;21(22):3006–3016. doi: 10.1101/gad.1575307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morley JF, Brignull HR, Weyers JJ, Morimoto RI. The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2002;99(16):10417–10422. doi: 10.1073/pnas.152161099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.David DC, et al. Widespread protein aggregation as an inherent part of aging in C. elegans. PLoS Biol. 2010;8(8):e1000450. doi: 10.1371/journal.pbio.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Link CD, Johnson CJ. Reporter transgenes for study of oxidant stress in Caenorhabditis elegans. Methods Enzymol. 2002;353:497–505. doi: 10.1016/s0076-6879(02)53072-x. [DOI] [PubMed] [Google Scholar]
- 35.Kahn NW, Rea SL, Moyle S, Kell A, Johnson TE. Proteasomal dysfunction activates the transcription factor SKN-1 and produces a selective oxidative-stress response in Caenorhabditis elegans. Biochem J. 2008;409(1):205–213. doi: 10.1042/BJ20070521. [DOI] [PubMed] [Google Scholar]
- 36.Lin K, Hsin H, Libina N, Kenyon C. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat Genet. 2001;28(2):139–145. doi: 10.1038/88850. [DOI] [PubMed] [Google Scholar]
- 37.Blake DJ, Weir A, Newey SE, Davies KE. Function and genetics of dystrophin and dystrophin-related proteins in muscle. Physiol Rev. 2002;82(2):291–329. doi: 10.1152/physrev.00028.2001. [DOI] [PubMed] [Google Scholar]
- 38.Gidalevitz T, Krupinski T, Garcia S, Morimoto RI. Destabilizing protein polymorphisms in the genetic background direct phenotypic expression of mutant SOD1 toxicity. PLoS Genet. 2009;5(3):e1000399. doi: 10.1371/journal.pgen.1000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gehrig SM, et al. Hsp72 preserves muscle function and slows progression of severe muscular dystrophy. Nature. 2012;484(7394):394–398. doi: 10.1038/nature10980. [DOI] [PubMed] [Google Scholar]
- 40.Morimoto RI. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 2008;22(11):1427–1438. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swaggart KA, Heydemann A, Palmer AA, McNally EM. Distinct genetic regions modify specific muscle groups in muscular dystrophy. Physiol Genomics. 2011;43(1):24–31. doi: 10.1152/physiolgenomics.00172.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heydemann A, Huber JM, Demonbreun A, Hadhazy M, McNally EM. Genetic background influences muscular dystrophy. Neuromuscul Disord. 2005;15(9-10):601–609. doi: 10.1016/j.nmd.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 43.Demontis F, Perrimon N. FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell. 2010;143(5):813–825. doi: 10.1016/j.cell.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Oosten-Hawle P, Porter RS, Morimoto RI. Regulation of organismal proteostasis by transcellular chaperone signaling. Cell. 2013;153(6):1366–1378. doi: 10.1016/j.cell.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barton ER, Morris L, Musaro A, Rosenthal N, Sweeney HL. Muscle-specific expression of insulin-like growth factor I counters muscle decline in mdx mice. J Cell Biol. 2002;157(1):137–148. doi: 10.1083/jcb.200108071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abmayr S, Gregorevic P, Allen JM, Chamberlain JS. Phenotypic improvement of dystrophic muscles by rAAV/microdystrophin vectors is augmented by Igf1 codelivery. Mol Ther. 2005;12(3):441–450. doi: 10.1016/j.ymthe.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 47.Ljubicic V, et al. Chronic AMPK activation evokes the slow, oxidative myogenic program and triggers beneficial adaptations in mdx mouse skeletal muscle. Hum Mol Genet. 2011;20(17):3478–3493. doi: 10.1093/hmg/ddr265. [DOI] [PubMed] [Google Scholar]
- 48.Chen CC, et al. FoxOs inhibit mTORC1 and activate Akt by inducing the expression of Sestrin3 and Rictor. Dev Cell. 2010;18(4):592–604. doi: 10.1016/j.devcel.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eghtesad S, Jhunjhunwala S, Little SR, Clemens PR. Rapamycin ameliorates dystrophic phenotype in mdx mouse skeletal muscle. Mol Med. 2011;17(9-10):917–924. doi: 10.2119/molmed.2010.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chamberlain JS. Duchenne muscular dystrophy models show their age. Cell. 2010;143(7):1040–1042. doi: 10.1016/j.cell.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sacco A, et al. Short telomeres and stem cell exhaustion model Duchenne muscular dystrophy in mdx/mTR mice. Cell. 2010;143(7):1059–1071. doi: 10.1016/j.cell.2010.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.