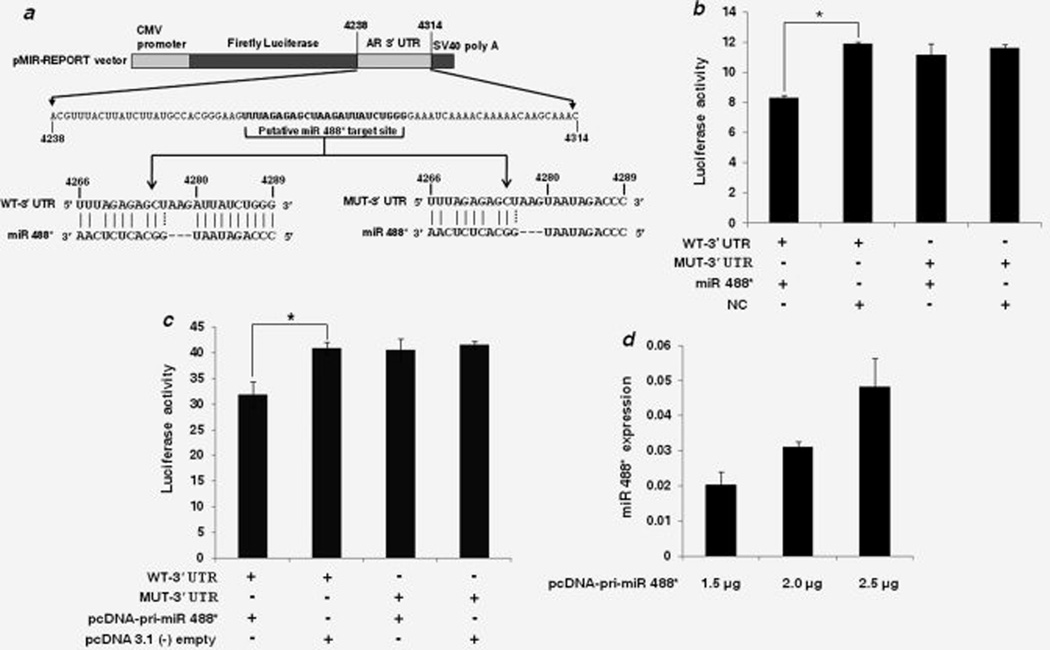
Figure 2.
AR is a direct target of miR 488*. (a) Schematic representation of firefly luciferase reporter construct containing 77 nucleotide sequence from AR 3’ UTR with either WT or mutant (MUT) miR 488* target site. In the MUT-3’ UTR construct, 10 nucleotides (4280–4289) in the seed matching region of the target site were mutated to their complementary nucleotides to disrupt miR 488* binding. (b) Luciferase reporter assay in CHO-K1 cells cotransfected with WT-3’ UTR or MUT-3’ UTR constructs and miR 488* mimic (10 nM) or NC mimic (10 nM) as indicated. (c) Luciferase reporter assay in CHO-K1 cells cotransfected with WT-3’ UTR or MUT-3’ UTR constructs and either pcDNA-pri-miR 488* construct (2.5 µg) or empty pcDNA 3.1(−) plasmid (2.5 µg) as indicated. (b and c) Luciferase activity is plotted as ratio of firefly to renilla luciferase activity. Each bar represents mean ± SE of three independent experiments. Asterisks indicate statistical significance as determined using the independent samples t-test (*p < 0.05). (d) miR 488* expression as determined by qRT-PCR. CHO-K1 cells were transfected with different amounts of pcDNA-pri-miR 488* construct as indicated and mature miR 488* expression was measured 48 hr post transfection. miR 488* expression is plotted as 2−ΔCt after normalization to snoRNA202 expression. Each bar represents mean ± SE of three independent experiments.