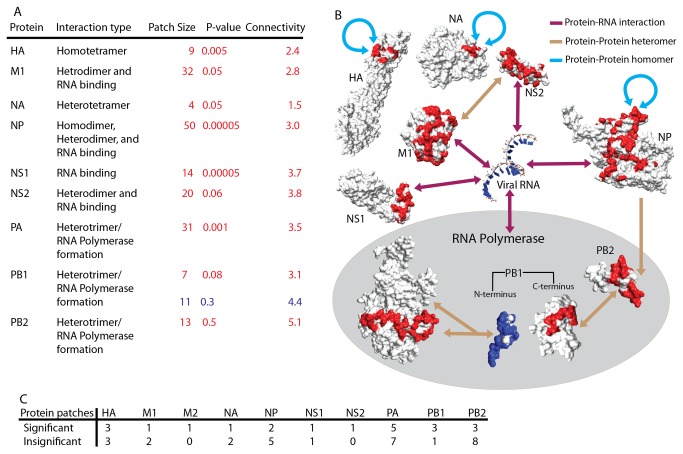
Figure 2. Conserved regions are exclusively associated with known intra-viral interaction positions.
A) Eight of the ten viral proteins have regions that are involved in known intra-viral interactions. For each interaction, we list the type of interaction, the size of the patch, En(x), and the patch connectivity. We determine En(x) as the expected number of randomly generated regions of a given size. We calculate the connectivity of the regions as the average number of neighbors each residue has in the patch. The color of the three right-most columns match to the color of the regions in panel B. B) Each of the eight proteins forms a unique interaction with (i) a copy of itself (indicated by a blue arrow), (ii) viral RNA (purple arrow), or (iii) another viral protein (tan arrow). Some conserved regions participate in more than one interaction. A uni-directional arrow indicates an interaction occurring between two proteins, but is not necessarily characterized by conserved regions on both proteins. The three proteins of RNA polymerase, PA, PB1, and PB2, are grouped by a grey oval. Shown is the interaction between the polymerase complex and the viral RNAs. C) The distribution of significant (En(x)≤0.05), marginal (0.05<En(x)≤0.1), and insignificant (En(x)>0.1) regions across all ten proteins.