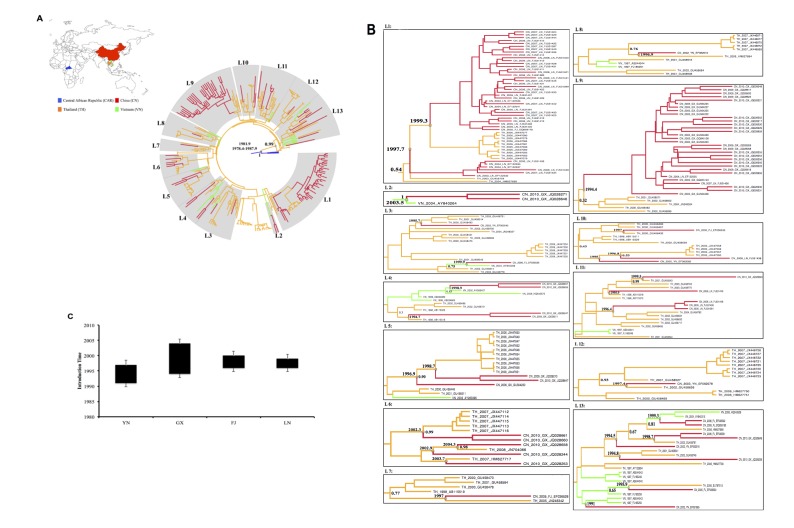
Figure 2. Time-scaled Bayesian Maximum Clade Credibility (MCC) tree.
A: Overall topology of MCC tree of the Chinese CRF01_AE lineages and the most closely related global sequences. Branches are colored according to the most probable location state of their descendent nodes. The color of the branches represents the geographic region from where the CRF01_AE sequences originated, according to the map given at the top left of the figure. The boxes highlight the position of the Chinese CRF01_AE clades. The first (CF) and second (TH) nodes representing initial nodes lineages to all lineages together with posterior probability values 1 and 0.99, respectively, are indicated. The median and 95% HPD interval of the tMRCA at the root of the tree is indicated. The tree was automatically rooted under the assumption of a relaxed molecular clock.
B: Close view of the Chinese CRF01_AE clades and the most closely related Asian sequences in MCC tree. The color of branches represents the geographic region from where the sequence originated. The names of CRF01_AE strains include reference to country origin, year of isolation, province (Chinese provinces) and GenBank Accession number. Countries represented are Central African Republic (CF), Thailand (TH), Vietnam (VN) and China (CN), the later represented by provincial sequences: Yunnan (YN), Guangxi (GX), Fujian (FJ) and Liaoning (LN). The posterior probability (PP) support values are indicated only at key nodes.
C: Estimated time-range of HIV-1 CRF01_AE earliest introductions into 4 border provinces (Yunnan (YN), Guangxi (GX), Fujian (FJ) and Liaoning (LN)) in China, the 95% highest posterior density (HPD) credible regions are provided as the range of state tMRCA.