Abstract
Background
Pipo was recently described as a new ORF encoded within the genome of the Potyviridae family members (PNAS 105:5897–5902, 2008). It is embedded within the P3 cistron and is translated in the +2 reading frame relative to the potyviral long ORF as the P3N-PIPO fusion protein. In this work, we first collected pipo nucleotide sequences available for different isolates of 48 Potyvirus species. Second, to determine the biological implications of variation in pipo length, we measured infectivity, viral accumulation, cell-to-cell and systemic movements for two Turnip mosaic virus (TuMV) variants with pipo alleles of different length in three different susceptible host species, and tested for differences between the two variants.
Results
In addition to inter-specific variation, there was high variation in the length of the PIPO protein among isolates within species (ranging from 1 to 89 amino acids). Furthermore, selection analyses on the P3 cistron did not account for the existence of stop codons in the pipo ORF, but showed that positive selection was significant in the overlapping region for Potato virus Y (PVY) and TuMV. In some cases, variability in length was associated with host species, geographic provenance and/or other strain features. We found significant empirical differences among the phenotypes associated with TuMV pipo alleles, though the magnitude and sign of the effects were host-dependent.
Conclusions
The combination of computational molecular evolution analyses and experiments stemming from these analyses provide clues about the selective pressures acting upon the different-length pipo alleles and show that variation in length may be maintained by host-driven selection.
Keywords: Bayesian phylogenetic methods, Host-range determinants, Molecular evolution, Potyvirus, Virus evolution, Virus fitness components
Background
The Potyviridae are one of the largest plant virus families, and many of its members represent major agricultural threats [1,2]. This family belongs to the picorna-like virus superfamily, for which overlapping ORFs have been described only in a few cases [3-5]. Until very recently, the genomic RNA of the Potyvirus genus, the largest genus within the Potyviridae, was thought to contain a single functional ORF encoding a polyprotein that is cleaved into 10 mature proteins (P1, HC-Pro, P3, 6 K1, CI, 6 K2, VPg, NIa-Pro, NIb, and CP) by three viral proteases: P1, HC-Pro and NIa-Pro [6]. However, Chung et al. [7] provided evidence of the existence of a new ORF in the reference sequences of 48 potyvirus species; and named it pipo (Pretty Interesting Potyviridae ORF). The pipo ORF is embedded within the P3 cistron and is translated in the +2 reading frame relative to the long ORF. Pipo is expressed as a fusion product with the N-terminal portion of P3 protein (ca. 25 kDa), P3N-PIPO, via ribosomal frameshifting or transcriptional slippage at a highly conserved motif at the 5′ end of pipo ORF [7]. Its length is quite variable among the different potyvirus species, ranging from 60 to 115 codons (inter-specific length variation) [7]. Wei et al.[8] and Wen & Hajimorad [9] have shown that P3N-PIPO plays a central role as a movement protein. This function is dependent on the interaction between P3N-PIPO and the host hydrophilic cation-binding protein PCaP1; and a knockout of the PCaP1 gene in Arabidopsis thaliana results in a severe reduction of TuMV accumulation and in milder symptoms [10]. Yeast two-hybrid experiments have shown that P3N-PIPO can interact with the small and large subunits of RubisCO, thereby potentially contributing to the development of symptoms [11]. Very recently, P3N-PIPO has been shown to be a virulence determinant that allows infection of plants carrying recessive resistance alleles [12]. However, the effects of genetic variation in pipo – in particular protein length – on interactions with host factors and the phenotypic consequences thereof are unknown.
Recently, during a computational survey of PVY variability [13], we made the fortuitous observation that pipo length was variable among isolates, with most isolates encoding for PIPO polypeptides of 73–76 amino acids long and one isolate encoding for a much shorter polypeptide of only 62 amino acids. These findings motivate the present study, which is organized in two parts. In the first part, we extended the in silico analysis of intra-specific variation in pipo length to the 48 potyvirus species used in the seminal study of Chung et al. [7]. We first identified those viruses with the largest numbers of isolates and alternative stop codons. For these viruses, we explored whether alternative stop codons may be under selection and whether differences in host species, geographic origin and/or other biological properties of the isolates may create different selective pressures that explain the prevalence of the different alleles in natural populations. The second part of the study is devoted to validating experimentally some of the previous in silico findings using two different length alleles of TuMV pipo. The first allele encodes the wild-type protein of 60 amino acids, hereafter referred to as pipo 61 . The second allele, dubbed pipo 70 , has the stop codons at positions 61 and 65 removed by site-directed mutagenesis, while retaining the stop codon at position 70. Thus, this variant encodes a longer protein consisting of 69 amino acids, which occurs in natural virus isolates. Both mutations at positions 61 and 65 were synonymous in the P3 reading frame. Then, we tested for allele-specific differences in infectivity, virus accumulation, and the speed of cell-to-cell and systemic movement in three host species: Nicotiana benthamiana, Brassica rapa and A. thaliana.
Methods
Alignment and recombination analysis
All available coding sequences that included the P3 cistron were retrieved from GenBank (48 virus species). Nucleotide sequences were translated and amino acid sequences were aligned with MUSCLE [14] as implemented in MEGA 5 [15]. Finally, the pipo ORF was located and its size was assigned for each isolate according to the position of the first stop codon in the +2 reading frame. Global information for the procedure is provided in Additional file 1: Table S1. For each intra-specific dataset showing differences in pipo length, the variables host, geographical origin and strain were retrieved whenever available (Additional file 1: Tables S2-S9). As a previous step to any phylogenetic or selection analysis, we performed recombination-detection analyses for the three largest datasets (Plum pox virus (PPV), PVY and TuMV) using RDP3 version 3.42 [16] to remove possible mosaic sequences, a common feature among the potyviruses [17-19]. RDP3 was used with the default configuration, except for the option of linear sequence and of disentangling overlapping signals. Only those breakpoints detected by at least three out of the eight methods implemented in RDP3 were accepted. No recombinant sequences were detected.
Phylogenetic and selection analyses
To check the strength of selection at the codon level, we estimated the differences in nonsynonymous and synonymous substitutions rates per site, d N – d S , using the fixed-effects likelihood (FEL) and internal branches fixed-effects likelihood (IFEL) [20,21]. These methods are implemented in http://www.datamonkey.org[22].
Phylogenetic analyses were performed using the GTR + Γ4 + I substitution model in a Bayesian MCMC framework, as implemented in BEAST version 1.7 [23]. Substitution rates were estimated using the relaxed uncorrelated exponential clock model. The MCMC was run for 108 generations to ensure convergence of all parameters. The inspection of posterior distributions and the estimation of the relevant evolutionary parameters were done with TRACER version 1.5 (tree.bio.ed.ac.uk/software/tracer). The first 10% of sampled trees were discarded as burn-in. Statistical significance of parameters was evaluated using the 95% highest probability density (HPD). In order to estimate the maximum clade credibility (MCC) phylogeny, including its posterior probabilities, the posterior set of trees (showing branch lengths in substitutions) obtained in BEAST was used. To do so, TREEANNOTATOR version 1.6.1 (beast.bio.ed.ac.uk) was used with 10% of the trees discarded as burn-in. The reliability of the MCC tree was evaluated using 95% HPD confidence intervals.
The CONTRAST program from PHYLIP version 3.69 (evolution.genetics.washington.edu/phylip.html), was used to detect correlations between pipo length and each of several biological factors while accounting for the underlying phylogeny. BATS version 1.0b2 [24] was used to elucidate the influence of host species, geographic origin and viral strain in the population structure. BATS computes the parsimony score PS[25], the association index AI[26] and the maximum monophyletic clade size MC[24] statistics, as well as assessing their significance. The first 10% of sampled trees were discarded as burn-in and 104 randomizations were performed to estimate the null distributions of the three statistics.
TuMV infectious clone
TuMV variants harboring different-length pipo alleles were generated by site directed mutagenesis on a plasmid containing TuMV cDNA (GenBank AF530055.2 with a few point mutations; isolate YC5 from calla lily [27]) tagged with GFP. The clone contains stop codons at position 61, 65 and 70 from pipo ORF sequence. The stop codon at position 61 was maintained in the wild-type allele pipo 61 . To generate the longer pipo 70 allele, we used the site-directed mutagenesis primers T3141G-PIPO65-2-S (5′-GGGGCAAGCAATGTGTGAAAAACGTACACTCTAG-3′) and T3141G-PIPO65-2-AS (5′-CTAGAGTGTACGTTTTTCACACATTGCTTGCCCC-3′) to replace stop codons at positions 61 and 65 by the most common codons among all the TuMV isolates, which at the same time only introduced synonymous mutations in the P3 ORF. All constructs were verified by DNA sequencing. DNA extraction was performed with the Qiagen Plasmid Maxi Kit (Qiagen) following the manufacturer’s instructions.
Measuring viral accumulation
To compare the accumulation of the TuMV-GFP encoding for different P3N-PIPO proteins, 56 four-week old N. benthamiana, 19 three-week old A. thaliana and 21 four-week old B. rapa plants were inoculated with DNA (approximately half of the plants with each pipo allele). Five μg of DNA (15 μL) and 5 μL of inoculation buffer (Carborundum 100 mg/mL, 50 mM potassium phosphate, pH 8.0) were mixed on one leaf per plant and gently rubbed. Inoculated plants were maintained in a growth chamber at cycles of 16 h of light at 25°C followed by 8 h of darkness at 22°C. In addition, control plants were inoculated with buffer and maintained in the same conditions.
GFP fluorescence was observed with a Leica MZ16F stereomicroscope, using a 0.5× objective lens, and GFP filters (Leica). Infected plants were randomly selected and harvested at different hours post-inoculation (hpi). Plants were ground into fine powder in a mortar with liquid N2, divided into aliquots and stored at -80°C. RNA extraction from 100 mg tissue per plant was performed using InviTrap® Spin Plant RNA Mini Kit (Invitek) following the manufacturer’s instructions. The concentration of total plant RNA extracts was adjusted to 50 ng/μL for each sample. Quantification of viral load was done by real time quantitative RT-PCR (RT-qPCR), using primers D359 (5′-CAATAGGTGCGAGAGAAGCACAC-3′) and D360 (5′-TAACCCCTGAACGCCCAGTAAG-3′). RT-qPCR reaction mix was prepared using the One-Step SYBR® Prime Script™ RT-PCR Kit II (Takara) with 100 ng RNA per reaction and following the instructions provided by the manufacturer. Amplifications were done using the StepOnePlus™ Sequence Analyzer 7500 (Applied Biosystems) with the following profile: 5 min at 42°C, 10 s at 95°C following 40 cycles of 5 s at 95°C and 34 s at 60°C. RT-qPCR reactions were performed in triplicate for each sample. Quantification results were further examined using SDS 7500 software v.2.2.2 (Applied Biosystems). Standards were prepared by linearization with BglII of the plasmid containing the TuMV infectious clone, followed by in vitro transcription using the mMessage mMachine® SP6 (Ambion) kit and following the manufacturer’s instructions. Known amounts of in vitro transcribed RNA were then added to a healthy plant total RNA preparation.
As a normalized measure of the rate of virus accumulation we used the Malthusian growth parameter, m, estimated as the slope of the linear regression of the log-transformed values of virus concentration to time (hpi). For statistical analyses, m values were fitted to a generalized linear model (GLM) with a Gamma distribution and a log-link function; “host” and “pipo” were treated as factorial random factors and “hpi” as a covariable in the model.
Minimum time necessary to trigger systemic infection
To study differences in the speed of colonization of systemic tissues of both TuMV pipo alleles, we followed the protocol described by Lafforgue et al. [28]. In short, ground infected tissue of N. benthamiana, obtained as described in the previous section, was re-suspended in inoculation buffer and the concentration of viral RNA was made equal for both genotypes. This sap was used to mechanically inoculate 158 four-week old N. benthamiana, 220 three-week old A. thaliana and 109 four-week old B. rapa plants (ca. half of the plants were inoculated with each pipo allele). Inoculated plants were maintained in a growth chamber under conditions of 16 h of light at 25°C and 8 h of darkness at 22°C. Inoculated leaves were removed at 24, 48, 72, 96, 120, and 144 hpi, and plants were maintained for up to two weeks under the same growth conditions. The effect of removing the inoculated leaf at different days post-inoculation (dpi) on the probability of systemic infection was determined by ascertaining whether there was GFP fluorescence in non-inoculated leaves after 9 dpi in the case of N. benthamiana and after 15 dpi for A. thaliana and B. rapa. GFP fluorescence was observed as described above. The frequency of systemically infected plants was fitted to a Binomial logistic regression equation using GLM; “host”, “pipo” and “dpi” (i.e., time at which inoculated leaves were removed) were treated as factorial random factors in the model.
Rate of expansion of primary infection foci
Four-week old N. benthamiana, three-week old A. thaliana and four-week old B. rapa plants were inoculated with ground infected tissue from N. benthamiana as described above. Inoculated leaves were examined at 48, 72 and 96 hpi with a Leica MZ16F stereomicroscope for formation of primary foci of infection. An individual photo was taken for each of many foci per leaf, and zoom was adjusted to the size of the focus and the intensity of fluorescent signal. All photos were normalized introducing the scale of amplification. The area (mm2) of a number of primary infection foci (103 from N. benthamiana, 45 from A. thaliana and 194 from B. rapa; ca. half for each of the two pipo alleles) was calculated using a MATLAB script, using the functions in the Image Processing Toolbox (MathWorks, Inc.). For statistical analyses, surfaces were divided by π and root-square transformed into effective radii (mm). The transformed data were fitted to a GLM with a Gamma distribution and log-link function; “host” and “pipo” were treated as factorial random factors and “hpi” as covariable in the model.
Results
Intra-specific variation in pipo length
Five or less isolates were available for 35 of the analyzed viruses, whereas more than 10 sequences were obtained only for eight viruses (Additional file 1: Table S1). Despite the reduced number of isolates available for most viruses, eight showed intra-specific alternative stop codons involving variable PIPO sizes (Table 1 and Additional file 1: Tables S2-S9). PVY showed the largest number of alternative stop codons (eight) among the 80 isolates included in the study. PPV and Papaya ringspot virus (PRSV) (with 69 and nine isolates, respectively) showed three alternative stop codons, whereas Pea seed-borne mosaic virus (PSbMV), Zucchini yellow mosaic virus (ZYMV), Sugarcane mosaic virus (SCMV), Potato virus A (PVA), and TuMV (with 3, 12, 13, 7, and 101 isolates, respectively) only presented two alternative stop codons each (Table 1). Whereas the minimal difference in PIPO size of only one amino acid was observed for ZYMV and SCMV, differences of six amino acids were found for PPV, nine amino acids for TuMV, and 11 amino acids for PRSV and PVA. Interestingly, the largest differences in size were observed for PVY (up to 31 amino acids) and especially for PSbMV, with three isolates having PIPO of 79 amino acids long and a fourth isolate having a protein as long as 168 amino acids. With respect to the frequency at which alternative stop codons were present in each virus, clear differences were also observed (Table 1), although it is worth mentioning that these frequencies do not take into account phylogenetic non-independence. A predominant stop codon was present in PPV, ZYMV, PVA, and TuMV. In the rest of viruses, however, alternative stop codons were present at similar frequencies, or at least no particular codon predominated.
Table 1.
Viruses showing alternative stop codons in the pipo ORF
| GenBank accession | Virus | PIPO size | # Isolates | Alternative stop codons | Frequency of functional |
|---|---|---|---|---|---|
|
NC_001445 |
Plum pox virus |
99 |
69 |
100 (1) |
0.014 |
| |
|
|
|
103 (59) |
0.87 |
| |
|
|
|
106 (68) |
0.116 |
|
NC_001616 |
Potato virus Y |
75 |
80 |
62 (1) |
0.0125 |
| |
|
|
|
65 (1) |
0.0125 |
| 73 (21) |
0.26 |
||||
| 74 (1) |
0.0125 |
||||
| 76 (58) |
0.64 |
||||
| 77 (79) |
0.05 |
||||
| 93 (80) |
0.0125 |
||||
|
NC_001671 |
Pea seed-borne mosaic virus |
79 |
3 |
80 (2) |
0.67 |
| 169 (1) |
0.33 |
||||
|
NC_001785 |
Papaya ringspot virus |
72 |
9 |
69 (1) |
0.11 |
| 73 (8) |
0.78 |
||||
| 80 (4) |
0.11 |
||||
|
NC_003224 |
Zucchini yellow mosaic virus |
76 |
12 |
76 (1) |
0.083 |
| 77 (12) |
0.917 |
||||
|
NC_003398 |
Sugarcane mosaic virus |
80 |
13 |
81 (9) |
0.69 |
| 82 (8) |
0.31 |
||||
|
NC_004039 |
Potato virus A |
83 |
7 |
84 (6) |
0.857 |
| 95 (7) |
0.143 |
||||
|
NC_002509 |
Turnip mosaic virus |
60 |
101 |
61 (87) |
0.861 |
| 70 (87) | 0.139 |
For each virus, the GenBank accession number of the reference sequence and its estimated PIPO size is given. The last three columns indicate the number of available isolates for each virus, the observed alternative stop codons (the number of isolates showing each stop codon is indicated within parenthesis) and the frequency at which a stop codon is the functional one for each virus, respectively. In the last column, frequencies are obtained for each virus by dividing the number of times a given stop codon appears first in the P3N-PIPO sequence by the total number of isolates.
Evaluating the strength of selection on alternative stop codons
Visual inspection allowed us to see that some changes in PIPO length resulted in nonsynonymous changes at the corresponding position of the P3 protein. This observation prompts the question of whether stop mutations were selected for their effects on P3N-PIPO or on P3. To answer this question, we ran selection analyses to evaluate the strength of selection on P3 for PPV, PVY and TuMV datasets, since they had the largest number of isolates. For PPV, no codons were found to be under positive selection in P3. For PVY, a single positively selected site was detected with FEL and IFEL methods at codon 179 in P3 (919 for the polyprotein), a site within the pipo ORF. A marginally significant positive selection (P = 0.052) was detected by FEL at codon 233 on P3 (amino acid 973 for the polyprotein). This codon is partially coincident with codon 62 from pipo, where a stop codon exists in one isolate (Table 1), besides showing a significant excess of non-synonymous changes at the corresponding positions of P3 protein. Taking these results into account, we constructed a contingency table classifying positively selected sites in P3 according to whether they occur in pipo or in the rest of P3 cistron. An excess of cases of positive selection within pipo was detected (P = 0.044). For TuMV, three positively selected sites were detected with the FEL method at codons 175, 180 and 203 on P3 (998, 1003 and 1026 for the polyprotein), all of them occurring in pipo. These three codons partially overlap with codons 11–12, 16–17, and 39–40, respectively, from pipo. An additional selected site outside pipo was also detected with the IFEL method at codon 238 on P3 (amino acid 1061 for the polyprotein). As previously observed for PVY, an excess of cases of positive selection within pipo was detected for TuMV (P = 0.035). In any case, the strength of selection on P3 seems not to account for the presence of alternative stop codons in PPV, PVY and TuMV pipo ORF.
Associations between pipo length and biological factors
Next, we sought for an association between the distribution of stop codons and three biological factors: host species, geographic origin and viral strain. In several cases, this association was obvious after visual inspection of the data. For PPV, the distribution of stop codons was related with the three biological factors (Additional file 1: Table S2). For instance, five out of the six isolates lacking stop codon 103 belonged to strain C while 47 out of the 48 isolates with this stop codon belonged to strain D. Also, it is noticeable that all isolates from cherry belonged to strain C and were European, which is in agreement with previous studies showing an association among host range, viral strain and geographic distribution [29,30]. Regarding PVY, in addition to the previously described association between isolates from strain C and the stop codon 77 [13], strain N isolates predominantly contained stop codon 73, while stop codon 76 was the most abundant among strain O isolates (Additional file 1: Table S3). For TuMV, a clear example of association is that stop codon 61 was present in all Asian isolates, whereas it was absent in some isolates from Europe, America and Oceania (Additional file 1: Table S4). Finally, SCMV stop codon 81 was present in all the maize isolates but absent in the sugarcane isolates (Additional file 1: Table S5).
All these associations were statistically significant (Fisher's exact test, P < 0.05 in all cases). However, it is necessary to recall that isolates from a virus do not represent independent observations but are phylogenetically related, thus jeopardizing the results from these simple association tests. Therefore, phylogenetic information has to be taken into account for proper statistical analyses. As before, we focused on PPV, PVY and TuMV data sets, since they encompassed large number of isolates. First, we wanted to test whether phylogenetic structure may explain the association between the distribution of stop codons and the three biological factors. For each virus, independent phylogenetic reconstructions were performed for the analysis of each factor, since the number of isolates for which information was available varied among factor. Phylogenetic analyses with the twelve data sets (four for each virus) were performed as described in the Methods section. After obtaining the MCC phylogenetic tree, BATS [24] was used to investigate the distribution of the three factors and of pipo length, along the phylogenies (as described in [13]). For PPV, these analyses showed that the three factors were not randomly distributed over the phylogenies but had a tendency to segregate on different clusters, and the same happened with the length of pipo (Table 2). To the extent of our knowledge, these analyses provide the first results concerning population structure and phylodynamics for PPV. For PVY and TuMV, significant associations were also observed (Tables 3 and 4, respectively), confirming previous results [13], although for PVY it was only marginally significant for the host species factor. Regarding TuMV, 15 different species were used for the analysis of the host factor. However, almost half of host species belong to the Solanaceae family (Table 4), and thus could not be treated as independent realizations of the trait. To solve this problem, we re-ran BATS but grouped isolates as belonging to the Brassicaceae (80 isolates) or the non-Brassicaceae (11 isolates). In this case, a significant association was observed, although it only arose because of the non-random distribution across the phylogeny of isolates from the Brassicaceae group (MC = 32.514, P = 0.011).
Table 2.
Association between geographic origin, host species, viral strain, and length of pipo allele with the distribution of viral isolates in the MCC phylogenetic tree obtained for PPV
| Analyses | # Isolates | Association statistics | Test value | P |
|---|---|---|---|---|
| Geographic origin |
|
PS |
22.722 |
< 0.001 |
| |
|
AI |
4.167 |
< 0.001 |
| Asia |
39 |
MC |
4.205 |
0.010 |
| America |
20 |
MC |
2.283 |
0.010 |
| Europe |
6 |
MC |
1.120 |
0.010 |
| Host species |
|
PS |
24.777 |
< 0.001 |
| |
|
AI |
4.614 |
< 0.001 |
| Apricot |
38 |
MC |
3.993 |
0.010 |
| Peach |
16 |
MC |
1.855 |
0.010 |
| Plum |
6 |
MC |
1.144 |
0.030 |
| Cherry |
4 |
MC |
1.054 |
0.010 |
| Almond |
1 |
MC |
NA1 |
|
| Strain |
|
PS |
5.850 |
< 0.001 |
| |
|
AI |
1.222 |
< 0.001 |
| D |
53 |
MC |
11.381 |
0.010 |
| C |
5 |
MC |
1.145 |
0.010 |
| M |
1 |
MC |
NA1 |
|
| PIPO length |
|
PS |
9.598 |
< 0.001 |
| |
|
AI |
2.070 |
< 0.001 |
| 103 |
59 |
MC |
9.409 |
0.010 |
| 106 |
9 |
MC |
1.345 |
0.010 |
| 100 | 1 | MC | NA1 |
AI: Association index, PS: Parsimony score, MC: Maximum monophyletic clade size.
1insufficient sample size (n < 2).
Table 3.
Association between geographic origin, host species, viral strain, and length of pipo allele with the distribution of viral isolates in the MCC phylogenetic tree obtained for PVY
| Analyses | # Isolates | Association statistics | Test value | P |
|---|---|---|---|---|
| Geographic origin |
|
PS |
36.260 |
< 0.001 |
| |
|
AI |
4.928 |
< 0.001 |
| Asia |
17 |
MC |
1.739 |
0.040 |
| Europe |
32 |
MC |
2.768 |
0.180 |
| America |
27 |
MC |
2.354 |
0.010 |
| Africa |
1 |
MC |
NA1 |
|
| Oceania |
1 |
MC |
NA1 |
|
| Host species |
|
PS |
7.922 |
0.040 |
| |
|
AI |
1.416 |
0.060 |
| Potato |
63 |
MC |
11.566 |
0.030 |
| Tobacco |
4 |
MC |
1.068 |
0.050 |
| Pepper |
1 |
MC |
NA1 |
|
| Black nightshade |
1 |
MC |
NA1 |
|
| Wild tomato |
1 |
MC |
NA1 |
|
| Tomato |
1 |
MC |
NA1 |
|
| Strain |
|
PS |
27.433 |
< 0.001 |
| |
|
AI |
4.092 |
< 0.001 |
| O |
46 |
MC |
4.361 |
0.010 |
| N |
23 |
MC |
2.148 |
0.010 |
| C |
8 |
MC |
1.120 |
0.010 |
| PIPO length |
|
PS |
27.323 |
< 0.001 |
| |
|
AI |
4.141 |
< 0.001 |
| 76 |
51 |
MC |
4.410 |
0.010 |
| 73 |
22 |
MC |
2.046 |
0.010 |
| 77 |
4 |
MC |
1.063 |
1 |
| 93 |
1 |
MC |
NA1 |
|
| 62 |
1 |
MC |
NA1 |
|
| 74 |
1 |
MC |
NA1 |
|
| 65 | 1 | MC | NA1 |
AI: Association index, PS: Parsimony score, MC: Maximum monophyletic clade size.
1insufficient sample size (n < 2).
Table 4.
Association between geographic origin, host species, viral strain, and length of pipo allele with the distribution of viral isolates in the MCC phylogenetic tree obtained for TuMV
| Analyses | # Isolates | Association statistics | Test value | P |
|---|---|---|---|---|
| Geographic origin |
|
PS |
30.362 |
< 0.001 |
| |
|
AI |
5.394 |
< 0.001 |
| Asia |
67 |
MC |
5.698 |
< 0.001 |
| Europe |
28 |
MC |
2.298 |
< 0.001 |
| America |
5 |
MC |
1.066 |
< 0.001 |
| Africa |
1 |
MC |
NA1 |
|
| Oceania |
1 |
MC |
NA1 |
|
| Host species |
|
PS |
44.461 |
< 0.001 |
| |
|
AI |
7.139 |
< 0.001 |
| Raphanus sativus |
45 |
MC |
3.662 |
0.090 |
| Brassica oleraceae |
12 |
MC |
1.372 |
0.200 |
| Brassica napus |
7 |
MC |
1.157 |
0.020 |
| Brassica campestris |
5 |
MC |
1.084 |
1 |
| Brassica pekinensis |
5 |
MC |
1.044 |
0.040 |
| Armoracia rusticana |
3 |
MC |
1.024 |
1 |
| Anemone coronaria |
2 |
MC |
1.008 |
1 |
| Calendula officinalis |
2 |
MC |
1.004 |
1 |
| Lactuca sativa |
2 |
MC |
1 |
1 |
| Allium ampeloprasum |
1 |
MC |
NA1 |
|
| Alliaria officinalis |
1 |
MC |
NA1 |
|
| Eustoma russellianum |
1 |
MC |
NA1 |
|
| Ranunculus asiaticus |
1 |
MC |
NA1 |
|
| Sisymbrium loeselli |
1 |
MC |
NA1 |
|
| Zaterdeschia spp. |
1 |
MC |
NA1 |
|
| Strain |
|
PS |
28.165 |
< 0.001 |
| |
|
AI |
4.894 |
< 0.001 |
| BR |
28 |
MC |
4.855 |
0.010 |
| B |
29 |
MC |
2.522 |
0.010 |
| (B) |
2 |
MC |
2.522 |
1 |
| B(R) |
1 |
MC |
NA1 |
|
| PIPO length |
|
PS |
13.392 |
< 0.001 |
| |
|
AI |
2.598 |
< 0.001 |
| 61 |
87 |
MC |
11.401 |
0.010 |
| 70 | 87 | MC | 1.475 | 0.010 |
PS: Parsimony score, AI: Association index, MC: Maximum monophyletic clade size.
1insufficient sample size (n < 2).
Next we used Felsenstein’s independent contrasts method [31] to evaluate the association between the distribution of stop codons and each of the three biological factors for these three viruses while continuing to take the underlying phylogenies into consideration. No significant correlations were observed for any of the viruses (correlation coefficients ranging between -0.050 and 0.120, and P > 0.33 in all cases).
Pipo length affects different fitness components of TuMV
As discussed in the previous section, Tables 2, 3 and 4 show that the phylogenetic distribution of PPV, PVY and TuMV isolates can be explained by the host species as well as by the length of pipo, suggesting that the apparent association between both traits may merely result from the phylogenetic relationships between isolates. To experimentally test whether pipo length had an effect on virus infection in different host species, we evaluated four different fitness components for both TuMV strains carrying the two pipo alleles of different lengths found for this virus (Table 1), pipo 61 and pipo 70 , across three alternative host species belonging to two botanical families, the Solanaceae (N. benthamiana) and the Brassicaceae (B. rapa and A. thaliana).
The first fitness component analyzed was infectivity, that is, the probability of establishing an infection upon inoculation of a given viral dose [32,33]. Consistent with the role of P3N-PIPO as viral movement protein [8,9], no differences in infectivity were observed between the two alleles (data not shown).
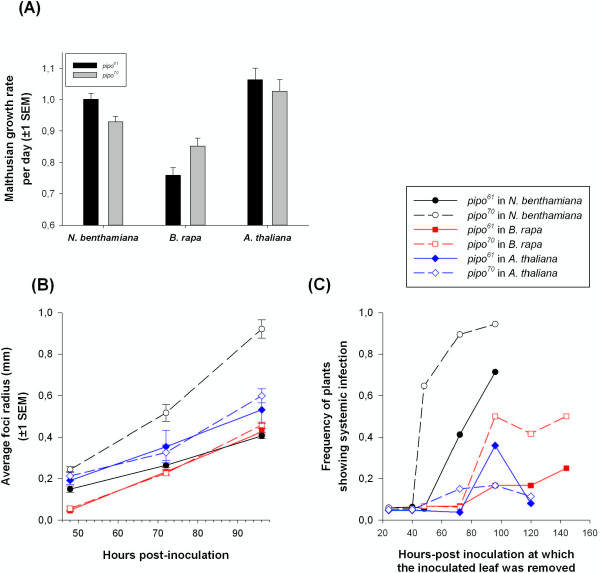
The second fitness component evaluated was Malthusian growth rate, which is a proxy for the rate of virus accumulation per day (Figure 1A). Significant differences in Malthusian growth rate exist between the three hosts (GLM test of “host” effect: χ2 = 70.8398, 2 d.f., P < 0.001), with growth rates being larger in A. thaliana and N. benthamiana and lower in B. rapa. More interestingly, a highly significant host-species-dependent effect of the pipo allele was found (GLM test of “host × pipo” interaction: χ2 = 13.994, 2 d.f., P = 0.001). TuMV-pipo 61 grew 7.8% faster in N. benthamiana and 3.6% faster in A. thaliana than the virus carrying the longer pipo 70 allele. In contrast, the virus carrying the pipo 70 allele grew 10.9% faster in B. rapa.
Figure 1.
Effects of two pipo alleles differing in length on TuMV biological properties. (A) Differences in accumulation rate between the wild-type pipo61 and the long pipo70 alleles across three experimental host species. Accumulation rate was measured as the per-day Malthusian growth parameter. (B) Rates of expansion of infection foci for the two alleles in the three experimental host species. (C) Speed of systemic infection for the two alleles in the three experimental host species. The time necessary to produce a systemic infection was measured as described in Lafforgue et al. [28]. Panels (B) and (C) share the same legend: solid symbols and lines correspond to the pipo61 allele, whereas open symbols and dashed lines to the pipo70 allele.
Next, we tested for differences between alleles in the rate of cell-to-cell movement by quantifying the average effective radius of expanding foci at increasing hpi, and comparing these dynamics across hosts (Figure 1B). Overall, no effect of the pipo alleles on the rate of foci expansion was detected (i.e., the slope of the regression lines; GLM test of “pipo × hpi” interaction: χ2 = 3.559, 2 d.f., P = 0.169). However, significant differences among slopes exist in the case of N. benthamiana plants (χ2 = 8.604, 1 d.f., P = 0.003), with TuMV-pipo 61 having a 2.55 (ratio of slopes) times faster cell-to-cell expansion than the virus carrying the long allele. No significant effects were observed in the other two hosts.
Finally, the fourth viral fitness component studied was the time required for the virus to exit the inoculated leaf and initiate a systemic infection. The faster a virus expands by cell-to-cell movement, the shorter the time necessary for the virus to reach the phloem and move upwards in the plant, following photo-assimilates towards sink tissues. Thus, in a set of plants this will translate into a higher frequency of systemic infections. To evaluate this, we followed the experimental protocol described by Lafforgue et al. [28], which consists of inoculating plants, removing the inoculated leaf after several hpi, and counting the number of plants developing systemic infection in each case. Figure 1C shows these data. As in the case of the cell-to-cell movement rate, no overall effect of the pipo allele was found (GLM test of the “pipo” main factor: χ2 = 0.003, 1 d.f., P = 0.956), although there was a significant three-way interaction (GLM test of the “host × pipo × hpi”: χ2 = 12.669, 2 d.f., P = 0.002). This interaction demonstrates that the time required for triggering a systemic infection indeed depends on the interaction between the host species and the pipo allele. At one extreme, systemic infection occurs sooner in N. benthamiana than in the other two hosts, and it is faster for the pipo 70 allele (Figure 1C). At the other extreme, time to establishing a systemic infection is longer in A. thaliana, with no significant differences among pipo alleles (Figure 1C). The case of B. rapa is intermediate among these two extremes, again with the longer allele promoting earlier systemic movement than the shorter one (Figure 1C).
Taken together, these results confirm that viral fitness depends on the interaction between the pipo allele and the host species being infected, thus providing an adaptive explanation for the different prevalence of pipo alleles among host species.
Discussion
The fact that pipo is embedded within P3 imposes strong functional and sequence constrains to Potyvirus evolution. The P3 cistron has been shown to be involved in adaptation to new hosts or overcoming resistance or tolerance in many potyviruses. This effect has been observed for Tobacco etch virus[34], TuMV [35-37], PSbMV [38], ZYMV [39,40], and Soybean mosaic virus (SMV) [41]. However, most of the virulence determinants described fall outside the P3N-PIPO protein. Interestingly, a key virulence determinant of SMV overlaps with both P3 and pipo ORFs, although it has been shown that these effects on virulence are due to the amino acid change in P3 but not in P3N-PIPO [42]. Also, a single mutation in the P3 cistron of TuMV that is essential for adaptation to Raphanus sativus also changes an amino acid in the P3N-PIPO protein [43], but the role of P3N-PIPO in this adaptation event remains to be determined. In any case, the fact that pipo is expressed as a fusion product with the N-terminal portion of P3 does not preclude from the potential influence of P3N-PIPO on virulence determinants mapped onto P3 and upstream P3N-PIPO. Since P3N-PIPO can also be involved in other functions [11], these potential multifunctional properties must be considered when analyzing particular positions in this protein.
Our results suggest that intra-specific variability in pipo size is a common feature in potyviruses. In this sense, five out of the eight viruses showing more than 10 isolates presented alternative stop codons, whereas this feature was only observed for three out of the 40 viruses for which less than 10 isolates were available. To date, experiments with the pipo ORF have consisted of introducing nonsynonymous mutations [44] or drastically premature stop codons [7,9]. However, experimental evidence of the potential effects of small differences in P3N-PIPO length, apart from those showed in the present work, is still missing. Consequently, further studies will be needed to understand whether the intra-specific variation in stop codon usage consists of neutral or nearly neutral polymorphisms that have been incorporated by drift events, or whether it plays a significant adaptive role in terms of host-range expansion, geographical dispersion and/or strain diversification.
Our experimental results confirm variability of the adaptive value of different pipo stop codons. We have not observed differences in infectivity, whereas we did observe differences in spread and accumulation. These observations suggest that the length of different alleles has no effect on initiating the infection. On the other hand, pipo length does have quantitative effects on virus spread, as might be expected for a mutation affecting the viral movement protein. Moreover, these effects are host dependent, thus further enhancing their potential selective value: certain alleles may be fitter in some hosts while other alleles may be so in alternative hosts.
Conclusions
Available information for the different poytivirus species suggests that intra-specific variability in pipo length is common. Our results also indicate that this variation may be maintained by host-driven selection, although further studies using other potyvirus models will be needed to shed some light on the generality of the results here shown.
Competing interests
The authors declared that they have no competing interests.
Authors’ contributions
JH and JMC performed the computational analyses, JH performed the experiments, SFE analyzed the empirical data, JMC and SFE conceived the study, JMC and SFE drafted the manuscript. All authors have read and approved the final manuscript.
Supplementary Material
Potyviruses analyzed in the present study. For each virus, the GenBank accession number of the reference sequence and its estimated PIPO size is given. The last two columns indicate the number of available isolates for each virus and the observed alternative stop codons, if present. The number of isolates showing a given stop codon is indicated in parentheses. Table S2. Available information for isolates belonging to Plum pox virus. The last three columns indicate the presence (+) or absence (-) of the alternative stop codons in each isolate. Table S3. Available information for isolates belonging to Potato virus Y. The last seven columns indicate the presence (+) or absence (-) of the alternative stop codons in each isolate. Table S4. Available information for isolates belonging to Turnip mosaic virus. The last two columns indicate the presence (+) or absence (-) of the alternative stop codons in each isolate. Table S5. Available information for isolates belonging to Sugarcane mosaic virus. The last two columns indicate the presence (+) or absence (-) of the alternative stop codons in each isolate. Table S6. Available information for isolates belonging to Pea seed-borne mosaic virus. The last two columns indicate the presence (+) or absence (-) of the alternative stop codons in each isolate. Table S7. Available information for isolates belonging to Papaya ringspot virus. The last three columns indicate the presence (+) or absence (-) of the alternative stop codons in each isolate. Table S8. Available information for isolates belonging to Zucchini yellow mosaic virus. Strain is not available for any of the isolates. The last two columns indicate the presence (+) or absence (-) of the alternative stop codons in each isolate. Table S9. Available information for isolates belonging to Potato virus A. The last two columns indicate the presence (+) or absence (-) of the alternative stop codons in each isolate.
Contributor Information
Julia Hillung, Email: juhil1@ibmcp.upv.es.
Santiago F Elena, Email: sfelena@ibmcp.upv.es.
José M Cuevas, Email: cuevast@uv.es.
Acknowledgements
We thank José A. Daròs for providing the TuMV-GFP infectious clone, Mario A. Fares and Guillaume Lafforgue for fruitful discussion, Guillermo Rodrigo for assistance with MATLAB and for critically reading of the manuscript, Mark P. Zwart for critically reading the manuscript, and Francisca de la Iglesia for excellent technical assistance. Funding for this work was provided by the Spanish Dirección General de Investigación Científica y Técnica grant BFU2012-30805 (to SFE), a predoctoral fellowship from the Spanish Ministerio de Economía y Competitividad (to JH), and a postdoctoral contract from CSIC JAE-doc program (to JMC).
References
- Riechmann JL, Lain S, García JA. Highlights and prospects of potyvirus molecular biology. J Gen Virol. 1992;73:1–16. doi: 10.1099/0022-1317-73-1-1. [DOI] [PubMed] [Google Scholar]
- Ward CW, Shukla DD. Taxonomy of potyviruses: current problems and some solutions. Intervirology. 1991;32:269–296. doi: 10.1159/000150211. [DOI] [PubMed] [Google Scholar]
- Kong WP, Roos RP. Alternative translation initiation site in the DA strain of Theiler's murine encephalomyelitis virus. J Virol. 1991;65:3395–3399. doi: 10.1128/jvi.65.6.3395-3399.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green KY, Ando T, Balayan MS, Berke T, Clarke IN, Estes MK, Matson DO, Nakata S, Neill JD, Studdert MJ. et al. Taxonomy of the caliciviruses. J Infect Dis. 2000;181:S322–S330. doi: 10.1086/315591. [DOI] [PubMed] [Google Scholar]
- van der Wilk F, Dullemans AM, Verbeek M, van den Heuvel JF. Nucleotide sequence and genomic organization of Acyrthosiphon pisum virus. Virology. 1997;238:353–362. doi: 10.1006/viro.1997.8835. [DOI] [PubMed] [Google Scholar]
- Urcuqui-Inchima S, Haenni AL, Bernardi F. Potyvirus proteins: a wealth of functions. Virus Res. 2001;74:157–175. doi: 10.1016/S0168-1702(01)00220-9. [DOI] [PubMed] [Google Scholar]
- Chung BY, Miller WA, Atkins JF, Firth AE. An overlapping essential gene in the Potyviridae. Proc Natl Acad Sci U S A. 2008;105:5897–5902. doi: 10.1073/pnas.0800468105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei T, Zhang C, Hong J, Xiong R, Kasschau KD, Zhou X, Carrington JC, Wang A. Formation of complexes at plasmodesmata for potyvirus intercellular movement is mediated by the viral protein P3N-PIPO. PLoS Pathog. 2010;6:e1000962. doi: 10.1371/journal.ppat.1000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen RH, Hajimorad MR. Mutational analysis of the putative PIPO of Soybean mosaic virus suggests disruption of PIPO protein impedes movement. Virology. 2010;400:1–7. doi: 10.1016/j.virol.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Vijayapalani P, Maeshima M, Nagasaki-Takekuchi N, Miller WA. Interaction of the trans-frame Potyvirus protein P3N-PIPO with host protein PCaP1 facilitates potyvirus movement. PLoS Pathog. 2012;8:e1002639. doi: 10.1371/journal.ppat.1002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Luo Z, Yan F, Lu Y, Zheng H, Chen J. Interaction between potyvirus P3 and ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO) of host plants. Virus Genes. 2011;43:90–92. doi: 10.1007/s11262-011-0596-6. [DOI] [PubMed] [Google Scholar]
- Choi SH, Hagiwara-Komoda Y, Nakahara KS, Atsumi G, Shimada R, Hisa Y, Naito S, Uyeda I. Quantitative and qualitative involvement of P3N-PIPO in overcoming recessive resistance against Clover yellow vein virus in pea carrying the cyv1 gene. J Virol. 2013;87:7326–7337. doi: 10.1128/JVI.00065-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas JM, Delaunay A, Visser JC, Bellstedt DU, Jacquot E, Elena SF. Phylogeography and molecular evolution of Potato virus Y. PLoS ONE. 2012;7:e37853. doi: 10.1371/journal.pone.0037853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics. 2010;26:2462–2463. doi: 10.1093/bioinformatics/btq467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilardi V, Nicola-Negri ED. Genetically engineered resistance to Plum pox virus infection in herbaceous and stone fruit hosts. GM Crops. 2011;2:24–33. doi: 10.4161/gmcr.2.1.15096. [DOI] [PubMed] [Google Scholar]
- Schubert J, Fomitcheva V, Sztangret-Wisniewska J. Differentiation of Potato virus Y strains using improved sets of diagnostic PCR-primers. J Virol Methods. 2007;140:66–74. doi: 10.1016/j.jviromet.2006.10.017. [DOI] [PubMed] [Google Scholar]
- Ohshima K, Tomitaka Y, Wood JT, Minematsu Y, Kajiyama H, Tomimura K, Gibbs AJ. Patterns of recombination in Turnip mosaic virus genomic sequences indicate hotspots of recombination. J Gen Virol. 2007;88:298–315. doi: 10.1099/vir.0.82335-0. [DOI] [PubMed] [Google Scholar]
- Pond SL, Frost SD, Grossman Z, Gravenor MB, Richman DD, Brown AJ. Adaptation to different human populations by HIV-1 revealed by codon-based analyses. PLoS Comput Biol. 2006;2:e62. doi: 10.1371/journal.pcbi.0020062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost SD. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol. 2005;22:1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- Delport W, Poon AF, Kosakovsky, Pond SL. Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics. 2010;26:2455–2457. doi: 10.1093/bioinformatics/btq429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J, Rambaut A, Pybus OG. Correlating viral phenotypes with phylogeny: accounting for phylogenetic uncertainty. Infect Genet Evol. 2008;8:239–246. doi: 10.1016/j.meegid.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Slatkin M, Maddison WP. A cladistic measure of gene flow inferred from the phylogenies of alleles. Genetics. 1989;123:603–613. doi: 10.1093/genetics/123.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TH, Donaldson YK, Brettle RP, Bell JE, Simmonds P. Identification of shared populations of Human immunodeficiency virus type 1 infecting microglia and tissue macrophages outside the central nervous system. J Virol. 2001;75:11686–11699. doi: 10.1128/JVI.75.23.11686-11699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Chao CH, Chen CC, Yeh SD, Tsai HT, Chang CA. Identification of Turnip mosaic virus isolates causing yellow stripe and spot on calla lily. Plant Dis. 2003;87:901–905. doi: 10.1094/PDIS.2003.87.8.901. [DOI] [PubMed] [Google Scholar]
- Lafforgue G, Tromas N, Elena SF, Zwart MP. Dynamics of the establishment of systemic Potyvirus infection: independent yet cumulative action of primary infection sites. J Virol. 2012;86:12912–12922. doi: 10.1128/JVI.02207-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs AJ, Ohshima K, Phillips MJ, Gibbs MJ. The prehistory of potyviruses: their initial radiation was during the dawn of agriculture. PLoS ONE. 2008;3:e2523. doi: 10.1371/journal.pone.0002523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasa M, Malinowski T, Predajna L, Pupola N, Dekena D, Michalczuk L, Candresse T. Sequence variability, recombination analysis, and specific detection of the W strain of Plum pox virus. Phytopathology. 2011;101:980–985. doi: 10.1094/PHYTO-12-10-0334. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Phylogenies and the comparative method. Am Nat. 1985;125:1–15. doi: 10.1086/284325. [DOI] [Google Scholar]
- Zwart MP, Daròs JA, Elena SF. One is enough: In vivo effective population size is dose-dependent for a plant RNA virus. PLoS Pathog. 2011;7:e1002122. doi: 10.1371/journal.ppat.1002122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart MP, Daròs JA, Elena SF. Effect of potyvirus effective population size in inoculated leaves on viral accumulation and the onset of symptoms. J Virol. 2012;86:9737–9747. doi: 10.1128/JVI.00909-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agudelo-Romero P, Carbonell P, Pérez-Amador MA, Elena SF. Virus adaptation by manipulation of host's gene expression. PLoS ONE. 2008;3:e2397. doi: 10.1371/journal.pone.0002397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner CE, Wang X, Tomimura K, Ohshima K, Ponz F, Walsh JA. The dual role of the potyvirus P3 protein of Turnip mosaic virus as a symptom and avirulence determinant in brassicas. Mol Plant Microbe Interact. 2003;16:777–784. doi: 10.1094/MPMI.2003.16.9.777. [DOI] [PubMed] [Google Scholar]
- Suehiro N, Natsuaki T, Watanabe T, Okuda S. An important determinant of the ability of Turnip mosaic virus to infect Brassica spp. and/or Raphanus sativus is in its P3 protein. J Gen Virol. 2004;85:2087–2098. doi: 10.1099/vir.0.79825-0. [DOI] [PubMed] [Google Scholar]
- Kim BM, Suehiro N, Natsuaki T, Inukai T, Masuta C. The P3 protein of Turnip mosaic virus can alone induce hypersensitive response-like cell death in Arabidopsis thaliana carrying TuNI. Mol Plant Microbe Interact. 2010;23:144–152. doi: 10.1094/MPMI-23-2-0144. [DOI] [PubMed] [Google Scholar]
- Hjulsager CK, Olsen BS, Jensen DM, Cordea MI, Krath BN, Johansen IE, Lund OS. Multiple determinants in the coding region of Pea seed-borne mosaic virus P3 are involved in virulence against sbm-2 resistance. Virology. 2006;355:52–61. doi: 10.1016/j.virol.2006.07.016. [DOI] [PubMed] [Google Scholar]
- Desbiez C, Gal-On A, Girard M, Wipf-Scheibel C, Lecoq H. Increase in Zucchini yellow mosaic virus symptom severity in tolerant zucchini cultivars is related to a point mutation in P3 protein and is associated with a loss of relative fitness on susceptible plants. Phytopathology. 2003;93:1478–1484. doi: 10.1094/PHYTO.2003.93.12.1478. [DOI] [PubMed] [Google Scholar]
- Glasa M, Svoboda J, Novakova S. Analysis of the molecular and biological variability of Zucchini yellow mosaic virus isolates from Slovakia and Czech Republic. Virus Genes. 2007;35:415–421. doi: 10.1007/s11262-007-0101-4. [DOI] [PubMed] [Google Scholar]
- Eggenberger AL, Hajimorad MR, Hill JH. Gain of virulence on Rsv1-genotype soybean by an avirulent Soybean mosaic virus requires concurrent mutations in both P3 and HC-Pro. Mol Plant Microbe Interact. 2008;21:931–936. doi: 10.1094/MPMI-21-7-0931. [DOI] [PubMed] [Google Scholar]
- Wen RH, Maroof MA, Hajimorad MR. Amino acid changes in P3, and not the overlapping PIPO-encoded protein, determine virulence of Soybean mosaic virus on functionally immune Rsv1-genotype soybean. Mol Plant Pathol. 2011;12:799–807. doi: 10.1111/j.1364-3703.2011.00714.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Z, Gibbs AJ, Tomitaka Y, Sánchez F, Ponz F, Ohshima K. Mutations in Turnip mosaic virus genomes that have adapted to Raphanus sativus. J Gen Virol. 2005;86:501–510. doi: 10.1099/vir.0.80540-0. [DOI] [PubMed] [Google Scholar]
- Choi IR, Horken KM, Stenger DC, French R. An internal RNA element in the P3 cistron of Wheat streak mosaic virus revealed by synonymous mutations that affect both movement and replication. J Gen Virol. 2005;86:2605–2614. doi: 10.1099/vir.0.81081-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Potyviruses analyzed in the present study. For each virus, the GenBank accession number of the reference sequence and its estimated PIPO size is given. The last two columns indicate the number of available isolates for each virus and the observed alternative stop codons, if present. The number of isolates showing a given stop codon is indicated in parentheses. Table S2. Available information for isolates belonging to Plum pox virus. The last three columns indicate the presence (+) or absence (-) of the alternative stop codons in each isolate. Table S3. Available information for isolates belonging to Potato virus Y. The last seven columns indicate the presence (+) or absence (-) of the alternative stop codons in each isolate. Table S4. Available information for isolates belonging to Turnip mosaic virus. The last two columns indicate the presence (+) or absence (-) of the alternative stop codons in each isolate. Table S5. Available information for isolates belonging to Sugarcane mosaic virus. The last two columns indicate the presence (+) or absence (-) of the alternative stop codons in each isolate. Table S6. Available information for isolates belonging to Pea seed-borne mosaic virus. The last two columns indicate the presence (+) or absence (-) of the alternative stop codons in each isolate. Table S7. Available information for isolates belonging to Papaya ringspot virus. The last three columns indicate the presence (+) or absence (-) of the alternative stop codons in each isolate. Table S8. Available information for isolates belonging to Zucchini yellow mosaic virus. Strain is not available for any of the isolates. The last two columns indicate the presence (+) or absence (-) of the alternative stop codons in each isolate. Table S9. Available information for isolates belonging to Potato virus A. The last two columns indicate the presence (+) or absence (-) of the alternative stop codons in each isolate.