Figure 4.
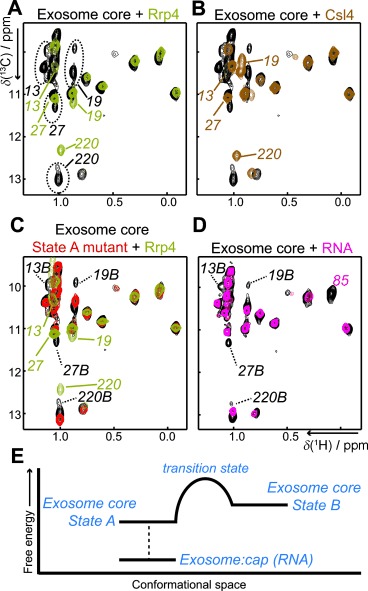
State A in the exosome core is stabilized upon interaction with cap proteins or with substrate RNA. (A, B) Spectra of the exosome core in the absence (black, dashed oval, two states) and presence of Rrp4 (A, olive) or Csl4 (B, brown). Upon cap interaction only a single state is observed in the exosome. The cap complexes interact with state A (see text), although this cannot be derived from the spectra directly due to the chemical shift changes induced by the cap. The signal to noise ratio for isoleucines 13, 19, 27, and 220 is larger than 10; thus, a potential minor state with a population of 25 % would have been observed in the spectra. C) Superposition of the WT exosome (black), the Rrp42 state A mutant where state B is no longer visible in the absence (red) and presence of Rrp4 (olive). In complex with Rrp4, Rrp42 is structured identically in the state A mutant exosome (olive) and in the WT exosome (Figure 1 A, olive spectrum) (Figure 4 A). D) Exosome core in the absence (black) and presence (pink) of an equimolar amount of RNA substrate (one RNA molecule per hexameric exosome complex). State B in the exosome core is no longer visible upon formation of the substrate–enzyme complex. In addition, residues close to the active site experience chemical shift perturbations (e.g Ile 85). E) Schematic representation of the reaction coordinates of the exosome core. States A and B have similar enthalpy; state A has a lower free energy due to increased entropy. The transition state is more disordered than the ground states A and B. State A interacts with the cap structure through a mechanism of conformational selection.
