Figure 1.
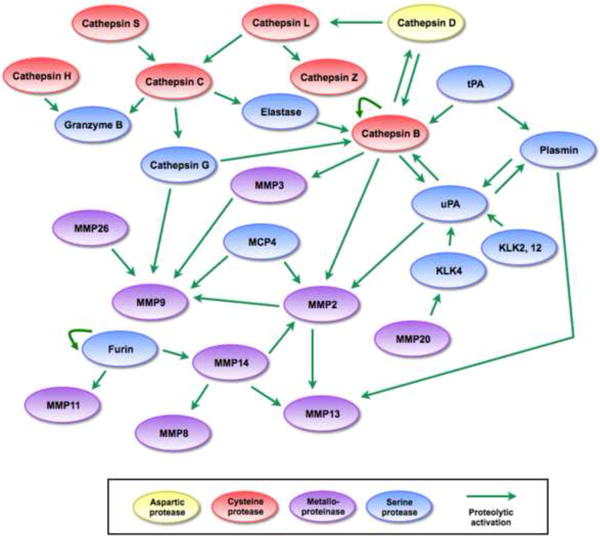
Different interactions within a proteolytic network.
Visualizing proteolytic interactions as a network of coordinated cascades reveals that the network has multiple entry points, interactions are multidirectional, and signals can be amplified in many directions. Examining protease-protease interactions specifically shows that similar to the cascade view, several proteases occupy nodes in the network and function as key regulators of proteolytic activity. Additionally, interactions involve proteases of different families and can proceed in multiple different pathways, allowing for some proteases to compensate for the absence of others. References for proteolytic interactions not described in the main text: cathepsin D activates cathepsin L [18], cathepsin L activates cathepsin Z [90], cathepsin B activates cathepsin D, MMP2, and MMP3 [18, 75], tissue-type plasminogen activator (tPA) activates cathepsin B and plasmin [91], MMP14 can activate MMP2 [92], MMP2, MMP14, and plasmin can activate MMP13 [93], MMP14 activates MMP8 [94], MMP26 can activate KLK4 [26], and furin activates MMP11 and MMP14 [31].
