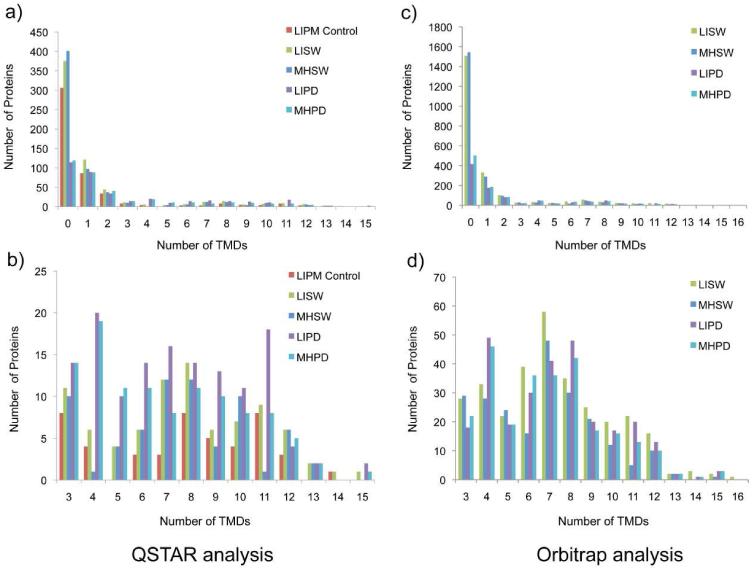
Figure 3. Histogram illustrations of the distributions of proteins with predicted TM helices identified from normal rat liver and hepatocellular carcinoma Morris hepatoma plasma membrane using different treatments.
Both data acquired by a QSTAR XL mass spectrometer (left) and an Orbitrap Velos mass spectometer (right) are shown for comparison. Identified proteins predicted with more than three TM helices are also shown in a more detailed view (below). Isolated plasma membranes were 1) LIPM Control: normal rat liver plasma membrane directly analyzed using the nonelectrophoretic gel-assisted digestion approach, 2) LISW or MHSW: LIPM or MHPM treated with carbonate washing, 3) LIPD or MHPD: LIPM or MHPM treated with carbonate washing followed by predigestion with trypsin. After the enhanced gel-assisted digestion, tryptic peptides were analyzed by LC-MS/MS for protein identification and quantification.