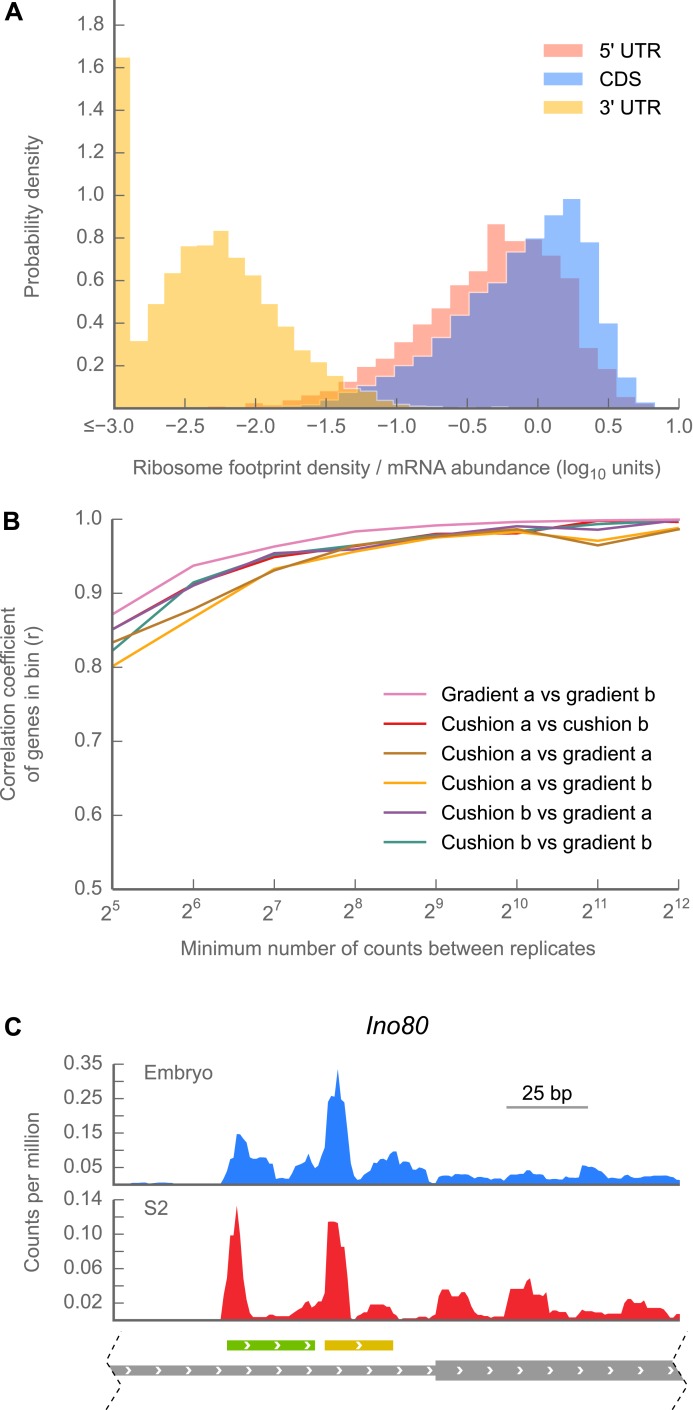
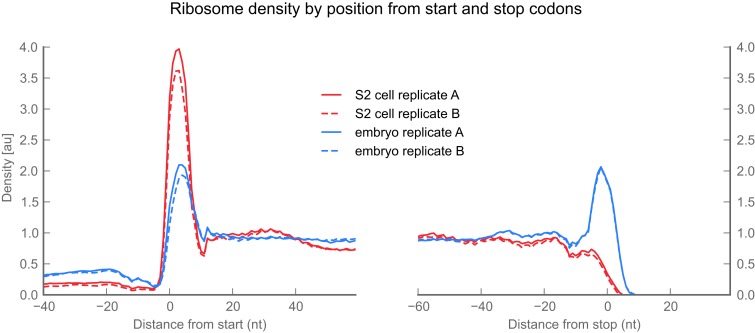
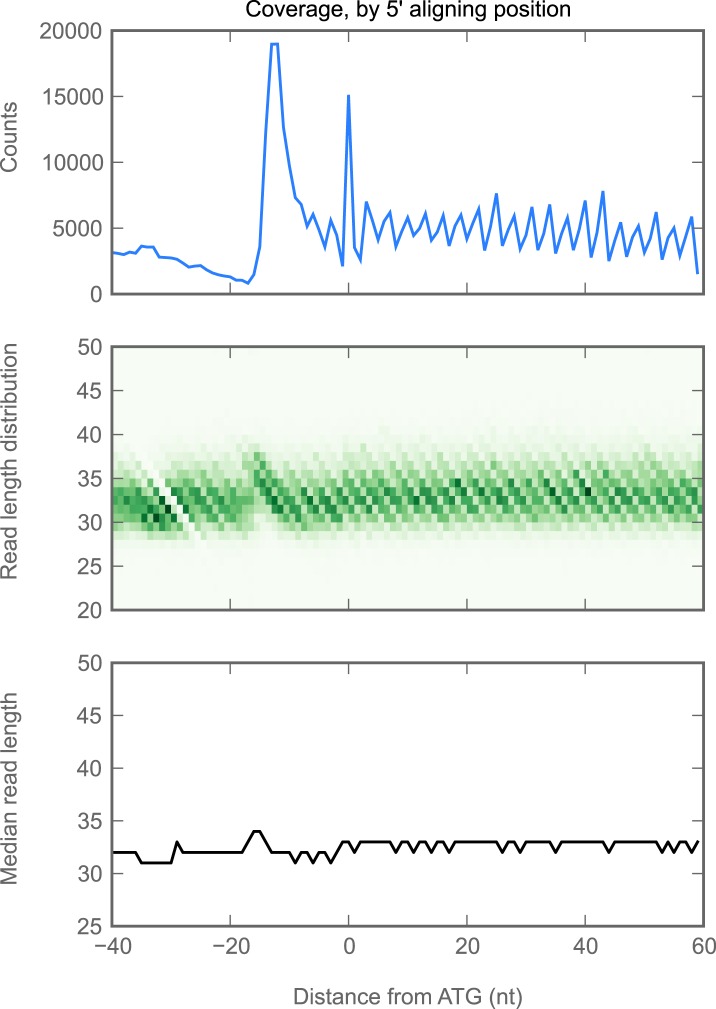
Figure 2. 5’ UTRs are translated.
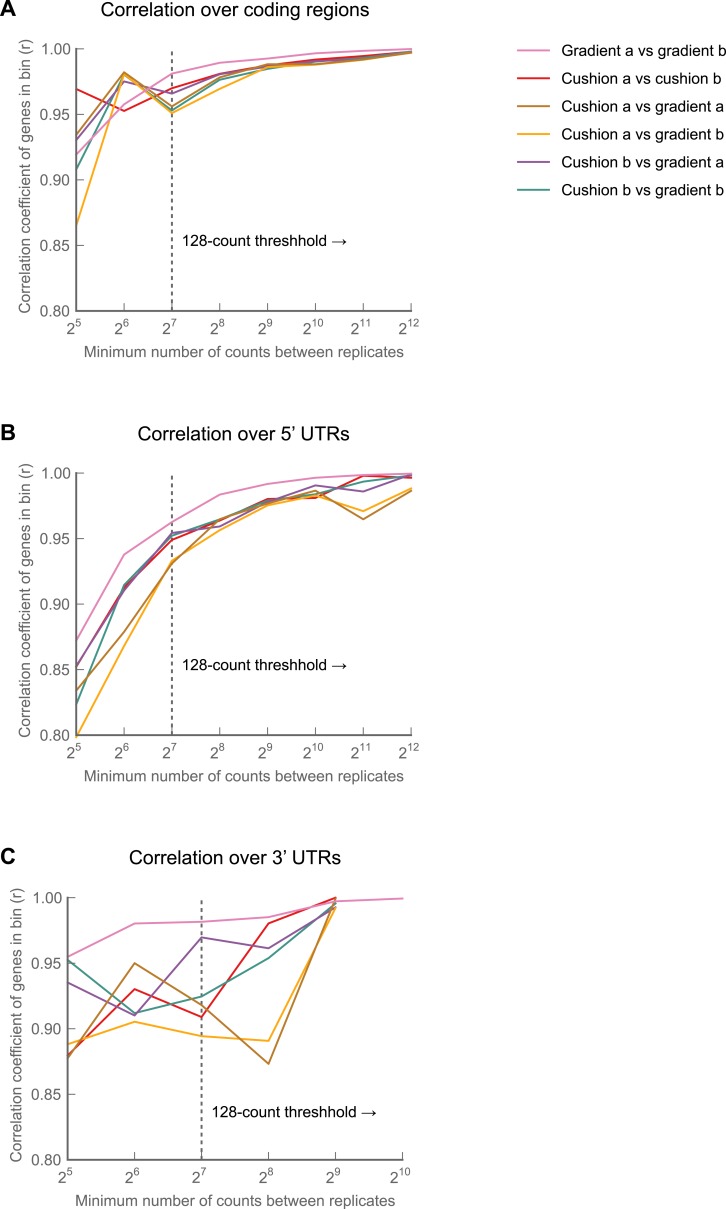
(A) Histograms of ribosome footprint density, corrected by mRNA abundance, for 5’ UTRs, coding regions (CDS), and 3’ UTRs in 0–2 hr embryos. (B) Measurements of ribosome footprint densities of 5’ UTRs agree comparably well across a range of sequencing depths, regardless of whether 80S monosomes are specifically isolated on a sucrose gradient or enriched in a cushion. For each pair of sequencing samples, Pearson correlation coefficients (r) of ribosome footprint density measurements for 5’ UTRs are plotted as a function of sequencing depth. (C) Example of ribosome density in 5’ UTRs corresponding to the locations of uORFs. Roughly ∼200 nt of the genomic locus Ino80 covering portions of the 5’ UTR (thin gray box) and CDS (thick gray box) are shown. In both 0–2 hr embryos and S2 cells, Initiation peaks are visible at the starts of uORFs starting with an ATG codon (green box) and a near-cognate TTG codon (yellow box) as well as at the annotated start codon (beginning of thick gray box). Source data for panels (A) and (B) may be found in supplementary table 1 (at Dryad: Dunn et al., 2013).