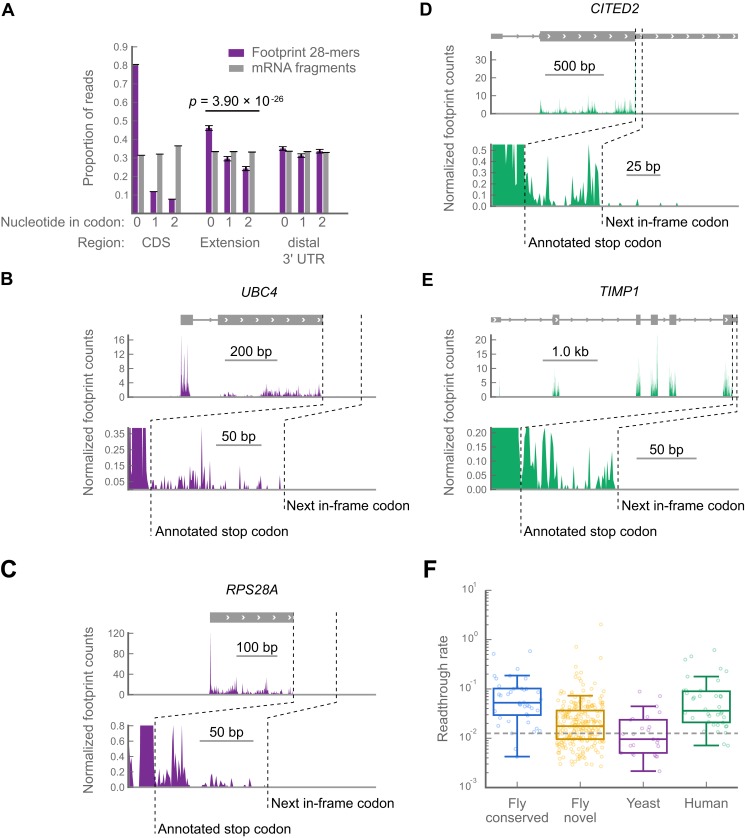
Figure 5. Readthrough occurs at specific stop codons in [psi-] yeast and in human foreskin fibroblasts.
(a) Triplet periodicity of 28-mers from yeast data in all non-overlapping coding regions (CDS), putative C-terminal extensions, and distal 3’ UTRs indicates that a signature of translation readthrough is visible in extensions on a bulk scale. Distal 3’ UTRs were estimated as 40 codon windows following putative extensions. Putative extensions and distal 3’ UTRs that overlap annotated coding regions, snoRNAs, snRNAs, tRNAs or 5’ UTRs were excluded from the analysis. (B and C) Examples of yeast transcripts that undergo readthrough, as in Figure 3B. (D and E) Examples of transcripts that undergo readthrough in human foreskin fibroblasts, as in Figure 3B. (F) Distribution of readthrough rates, by organism, for all extensions of sufficient length not to be covered by bleedthrough from termination peaks (‘Materials and methods’). Dashed line: fifth percentile of readthrough rate in conserved extensions in D. melanogaster, 1.2%. Source data may be found in supplementary tables 2, 3, and 4 (at Dryad: Dunn et al., 2013).