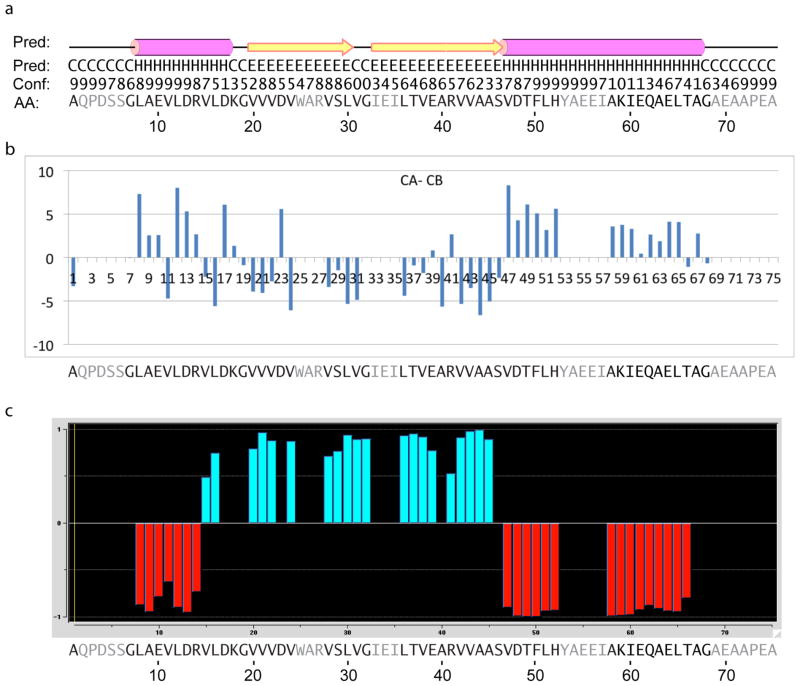
Fig. 9.
Secondary structure results for GvpA in H. salinarum. In all panels, the amino acid residues that remain unassigned are shaded in gray.
(a) Repeat of the PSIPRED prediction shown in fig. 3.
(b) Results of CA-CB analysis. Positive and negative values identify helix and strand conformations, respectively.
(c) Results of TALOS+ analysis. Negative and positive values identify helix and strand conformations, respectively.