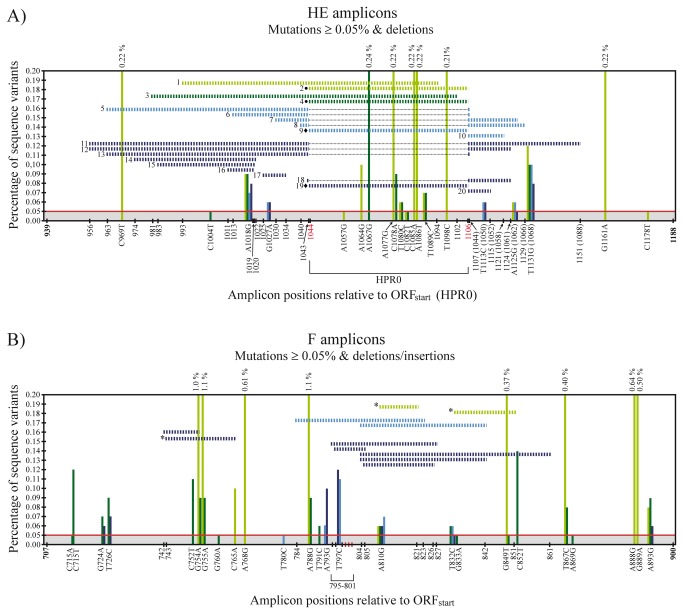
Figure 1. Position, percentage and type of single nucleotide variants (vertical lines), and deletion-insertions and deletions (horizontal lines) detected in reads from UDPS of HE gene (A) and F gene (B) amplicons from a non-virulent HPR0- and a virulent ISAV strain.
Grey shading and the red horizontal line illustrate that only mutations present in frequencies ≥ 0.05 % are included. Color coding; light green = single screening sample, dark green = pooled screening sample, light blue = single outbreak sample, dark blue = pooled outbreak sample. The three red vertical lines in B) is the L266/Q266 codon in the F gene. Numbering of deletion events in A) correspond to numbering in Figures 2a, b . * = deletion-insertion types observed in F reads (regions deleted are shown, see Table S3 for details), • = outbreak delHPRs in screening samples, and♦ = full-length HPRs in outbreak samples. Numbers in bold indicate ORFstart and ORFend positions in HPRO HE- and F genes not containing insertions. Red numbers indicate start and end of the full-length HPR, and numbers in parenthesis downstream of this region are the corresponding positions relative ORFstart in the outbreak delHPR. Dotted lines illustrate the full-length HPR portion not present in the delHPR.