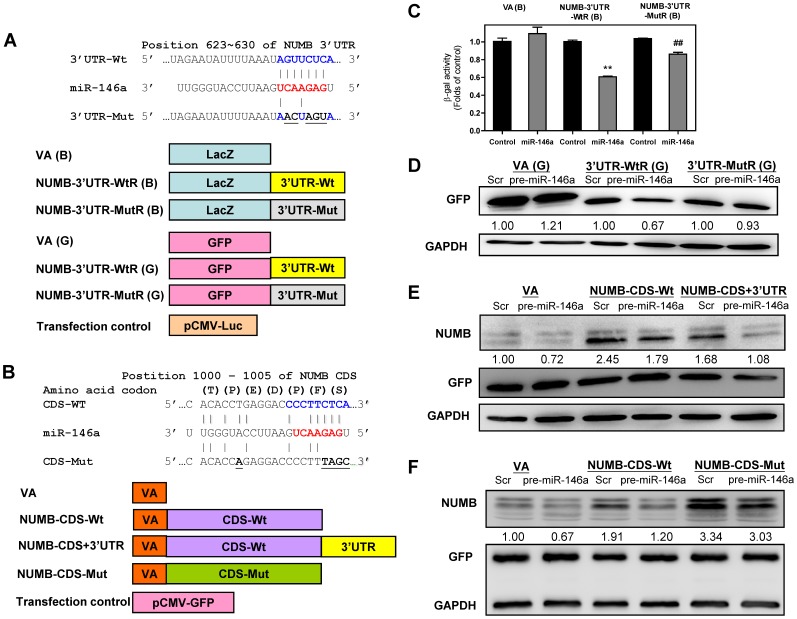
Figure 9. Validation of NUMB as a target for miR-146a.
(A, B) Schematic diagram of the reporter constructs and expression constructs, respectively. The vertical lines indicate the matching of the sequences. Wt, wild-type; Mut, mutation. CDS, coding sequence; 3′UTR, 3′untranslated region. The characters in parentheses are the one-letter codes for amino acids. The NUMB-CDS-Mut contains mutations affecting five nucleotides, but does not change the amino acid sequence of the protein. (C) Reporter activity assays using LacZ as the reporter gene in SAS cell subclones. Data shown are the means ± SE from triplicate analysis. **, p<0.01, comparison with controls; ##, p<0.01, comparison across WtR(B) and MutR(B); Mann-Whitney test. (D - F) Western blot analysis of the SAS cells transfected with the pre-miR-146a mimic, reporter constructs or expression plasmids. (D) GFP expression. SAS cells are transfected with pre-miR-146a mimic and GFP reporters. The analysis shows that the wild-type 3′UTR is associated with the repression of GFP expression, while the mutations in the matched sequences reverse the repression. (E, F) NUMB and GFP expression. GFP expression level is used as the transfection control. (E) This shows that the exogenous wild-type NUMB isoform 4 expression in the control cells is decreased in the presence of the 3′UTR, and is further decreased in cells that have exogenous miR-146a expression. (F) This shows that the mismatch mutations in the CDS of NUMB reverse the targeting effects of both the endogenous and the exogenous miR-146a. Scr, scramble; VA, vector alone. The pictures in (D – F) are representative pictures of two distinctive Western blot analysis. Numbers below gels depict the expression values for GFP normalized against GAPDH (in D) and for NUMB normalized against GFP (in E and F).