Figure 5.
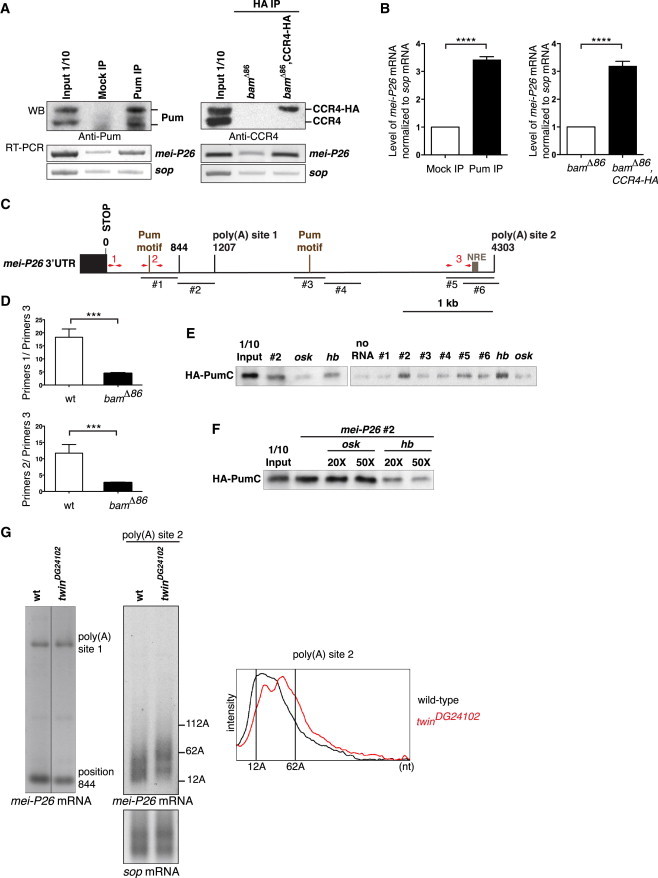
mei-P26 mRNA Is a Target of the Nos/Pum/CCR4 Complex in the GSCs
(A and B) RNA immunoprecipitations (IP) with the anti-Pum antibody in bamΔ86 ovarian extracts (left panels, Mock IP: preimmune serum) and with the anti-HA antibody in bamΔ86 and nos-Gal4, bamΔ86/UASp-CCR4-HA, bamΔ86 (bamΔ86, CCR4-HA) ovarian extracts (right panels). sop mRNA was used as a negative control. (A) Top panels show protein IP using western blots (WB). Bottom panels show the enrichment of mei-P26 mRNA in IP compared to in mock IP, visualized by RT-PCR. Inputs are protein or RNA extracts (1/10) prior to immunoprecipitation. Two (HA IP) and four (Pum IP) IPs were performed with similar results. (B) Quantification by qRT-PCR. Normalization was with sop mRNA. Quantifications were done in triplicate, and error bars represent SD. ∗∗∗∗p < 0.0001 using the t test.
(C) Schematic representation of mei-P26 3′ UTR showing position 844, the two poly(A) sites identified by RNA circularization and the three potential Pum binding motifs, including the NRE-type motif. RNA fragments tested in RNA pull-down assays (#1 to #6) and primer sets (1, 2, and 3) used to quantify the utilization of alternative poly(A) sites are also indicated. Coordinates are from the STOP codon, 1 being the first nucleotide of the 3′ UTR (see also Figure S6).
(D) Quantification by qRT-PCR of ratios of mRNA levels upstream of position 844 to just upstream of poly(A) site 2, in wild-type early ovarian stages and bamΔ86 mutant ovaries. Two primer sets (1 and 2) localized upstream of position 844 gave similar results. Means are from two independent RNA extracts quantified in triplicate, and error bars represent SD. ∗∗∗p < 0.0005 using the t test.
(E) RNA pull-down assays using the mei-P26 RNA fragments #1 to #6 shown in (C) and HA-PumC protein. Input is the in vitro synthesized protein (1/10) prior to RNA pull-down. osk and hb RNA fragments are negative and positive controls for Pum-C binding, respectively.
(F) RNA pull-down competition assays of mei-P26 RNA fragment #2 with increasing amounts of osk or hb unlabeled RNA fragments. Lane 2 is HA-PumC pull-down with fragment #2 in the absence of competitor. Input is as in (E).
(G) PAT assays of mei-P26 mRNA with specific primers allowing to measure potential poly(A) tails downstream of position 844 and poly(A) site 1 (left panel), and poly(A) site 2 (right panel), in wild-type and twinDG24102 early ovarian stages. sop was used as a control mRNA. PAT assay profiles of mei-P26 poly(A) site 2, using ImageJ, are shown.
