Figure 1.
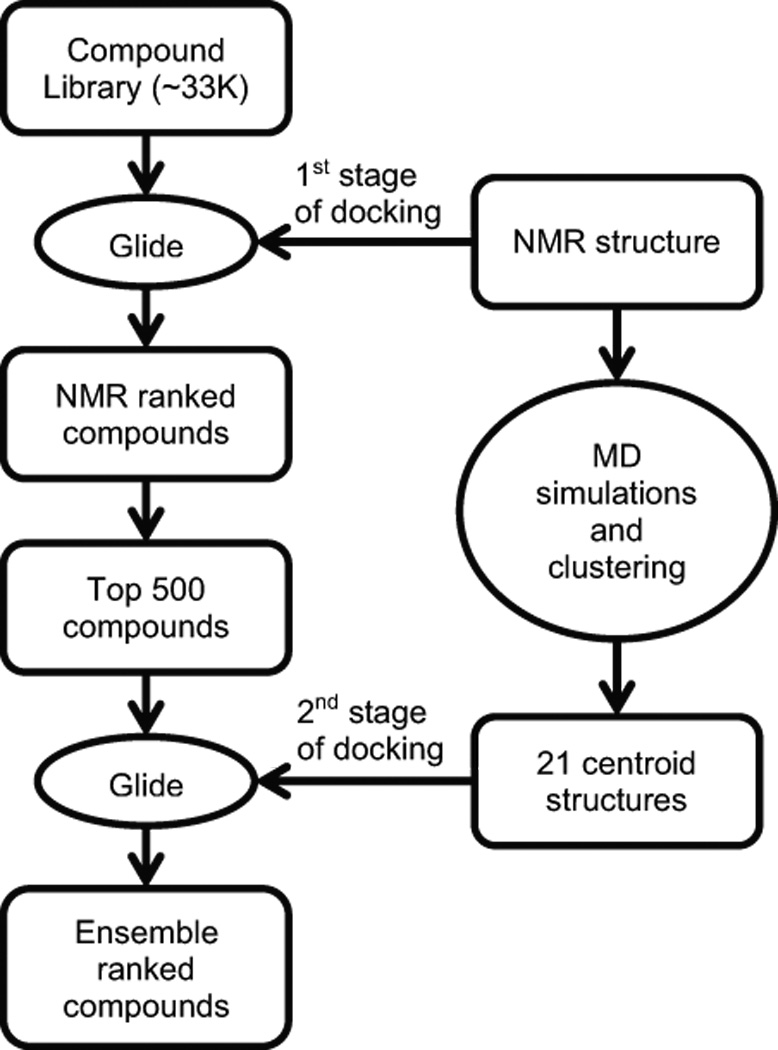
Overview of the two-staged virtual screening procedure that used the Relaxed Complex Scheme. In the first stage, small molecules from the ZINC compound library are docked using the program Glide to the NMR structure of substrate bound form of SrtA (PDB ID: 2KID). In the second stage, six 100 ns MD simulations of the NMR structure are performed and their snapshots are clustered by an RMSD-based algorithm, generating 21 clusters. The top 500 compounds obtained from the first screen are then docked using Glide to 21 centroid structures that represent each of the 21 clusters. Finally, the compounds are ranked by three different methods and the top 15 compounds in each ranking category are selected for experimental testing.
