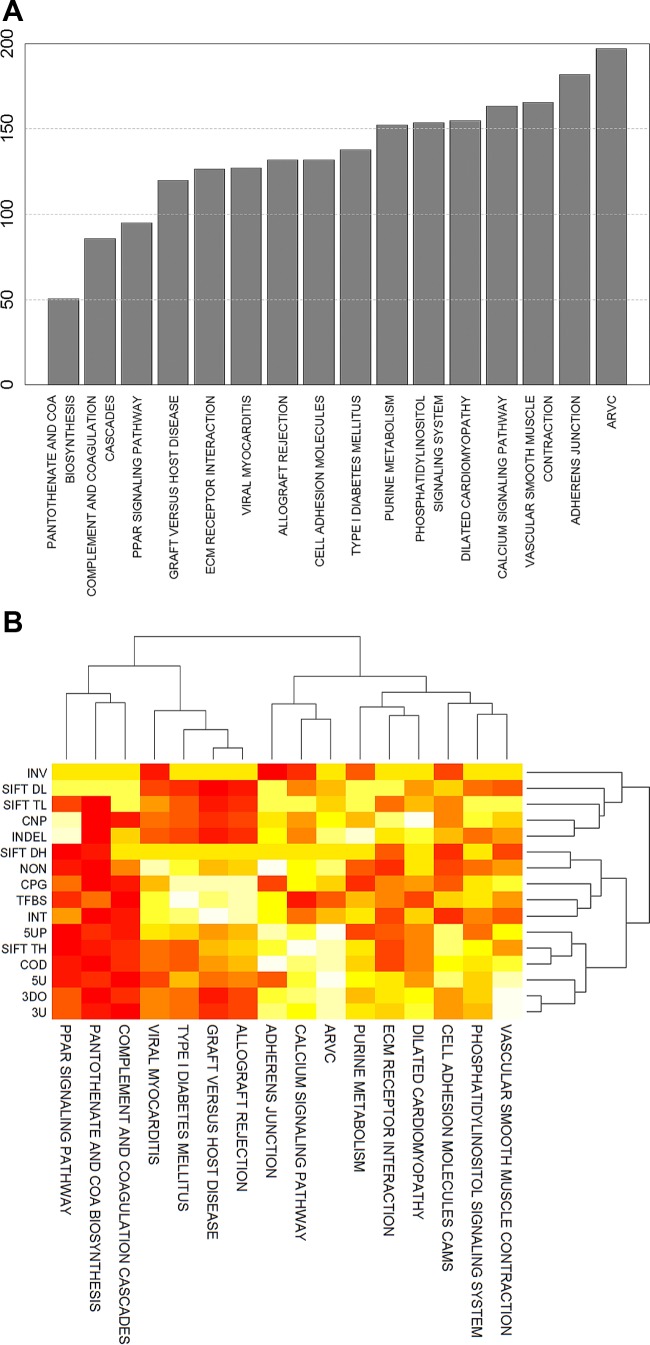
Fig. 6.
Ranking pathways by their predicted SNP burden. A: sixteen pathways identified as being significantly associated with ΔV̇o2max by both IGSEA4GWAS and GSA-SNP were scored for functional and positional effects of the SNPs contained in their gene members. Pathways were ranked on the basis of the sum of scores (adjusted for pathway size) such that pathways with lower ranks are those with higher predicted SNP burden. B: two-way hierarchical clustering of pathways and possible SNP effects on the basis of pathway scores in each category (white, lower scores; red, higher scores). Pathways that are more similar by type of SNP burden are clustered more closely compared with pathways that have different SNP burden profiles. SNP categories are abbreviated as follows: INV, inversion; SIFT DL, amino acid substitution (damaging, low confidence); SIFT TL, amino acid substitution (tolerated, low confidence); CNP, copy number polymorphism; INDEL, insertions and deletions; SIFT DH, amino acid substitution (damaging, high confidence); NON, noncoding region; CPG, CpG island region; TFBS, transcription factor binding sites; INT, intronic; 5UP, 5′-upstream (5 kb); SIFT TH, amino acid substitution (tolerated, high confidence); COD, coding region; 5U, 5′ untranslated region (5′UTR); 3DO, 3′-downstream (5 kb); 3U, 3′ untranslated region (3′UTR).