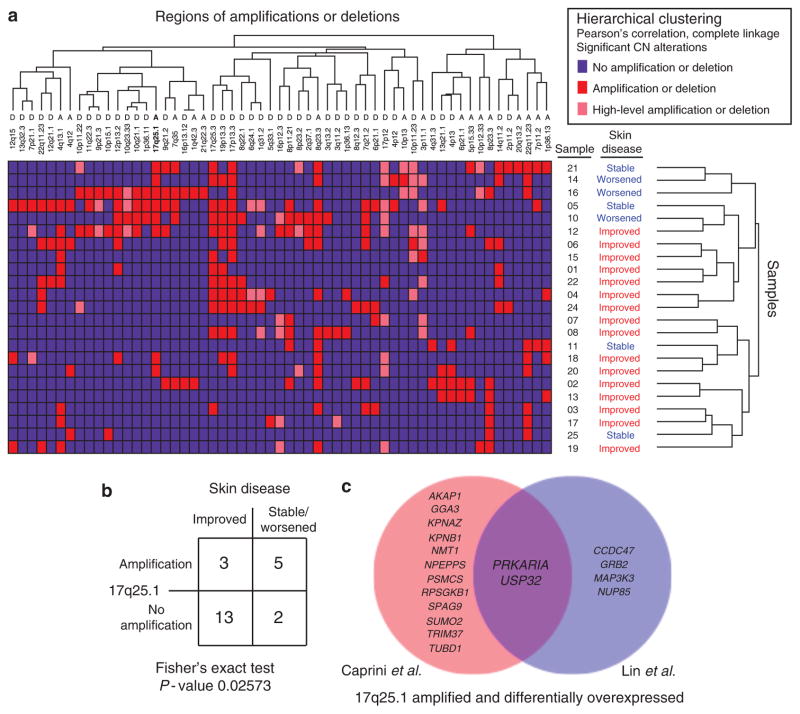
Figure 3. 17q25.1 amplification candidate targets.
(a) Unsupervised analysis of significant copy-number (CN) mutations in cutaneous T-cell lymphoma (CTCL) as defined by GISTIC (Genomic Identification of Significant Targets in Cancer) analysis was performed with hierarchical clustering. Each patient’s skin disease severity after 1 year of treatment is annotated next to the sample number. (b) Fisher’s exact test showed 17q25.1 amplification to be more common in patients with stable/worsened skin disease after 1 year of treatment; however, this was not significant after Bonferroni multiple-hypothesis correction. (c) Using Comparative Marker Selection, differential expression analysis of samples with 17q25.1 amplification versus no amplification reveals candidate targets that are amplified and overexpressed in the data set of Caprini et al. (2009; shown in red), our data set (shown in blue), and the overlap (shown in purple).