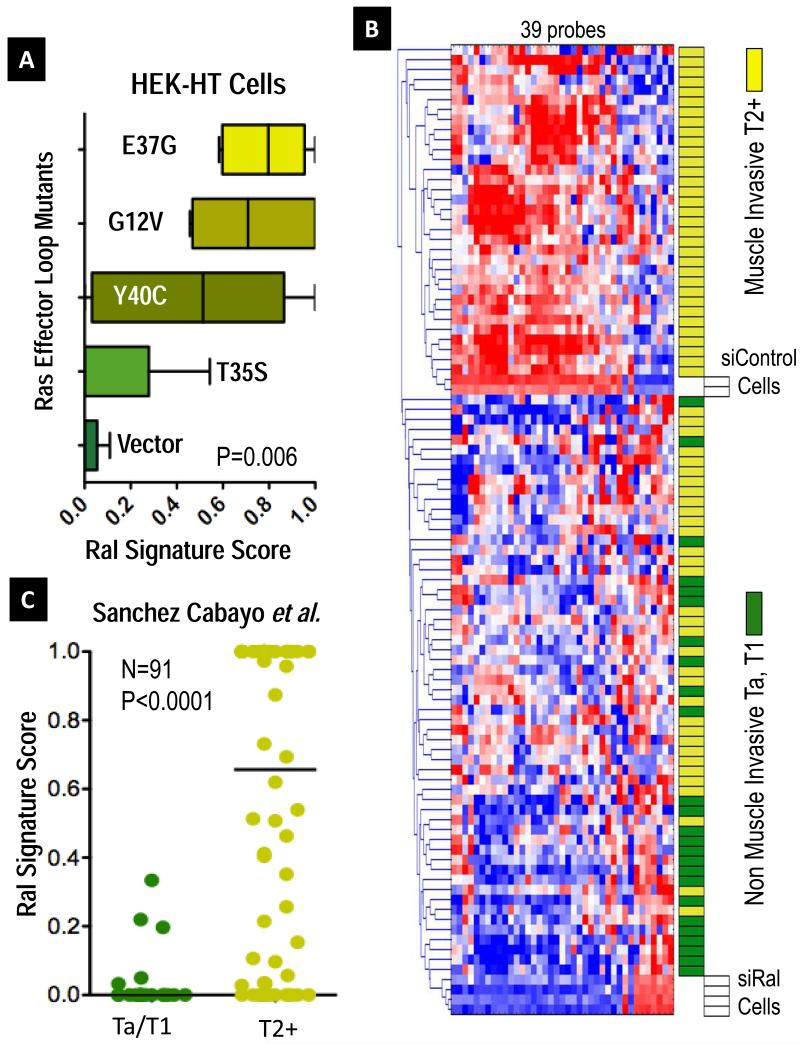
Figure 2. The Transcriptional Signature of Ral GTPases.
A Ral Transcriptional Signature consisting of 39 probes was developed from genes regulated 2-fold by RalA and RalB expression in UM-UC-3 cells. To validate the association of this signature with Ral across cell types, we used a weighted nearest neighbor (WNN) algorithm (21), which outputs a score from 0, Signature negative, to 1, Signature positive. We classified samples from published data from Chang et al. (26), where quintuplicate preparations of HEK-HT cells stably expressing vector control, G12V oncogenic HRAS, or indicated G12V HRAS effector loop mutants were profiled by oligonucleotide microarrays. A. Boxplots of median and range (whiskers) Ral signature scores for the indicated vector control or Ras mutant, finding higher scores in oncogenic Ras (G12V) and its effector loop mutant (G12V/E37G) stimulating the Ral pathway (P=0.006, Kruskal-Wallis test). B. Hierarchical cluster analysis of gene expression data for 91 bladder cancers (22) and control siRNA treated UM-UC-3 cells (siControl) or RalA and RalB-depleted siRNA duplexes (siRal), showing association of the signature with muscle-invasive tumors (yellow blocks). C. Expression of the Ral signature from cases in B. was again quantitated as before and dotplotted, medians indicated by lines. Differences in score distributions between non-muscle invasive pTa/T1 cases and muscle invasive pT2+ cases were tested by the Mann-Whitney test. Similar significant findings were identified in six additional cohorts comprising over 500 additional patients (see Figure S5A-F).