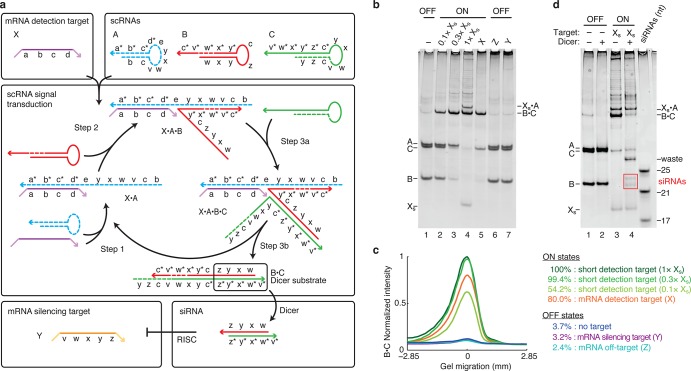
Figure 2.
Conditional catalytic DsiRNA formation using metastable scRNAs. (a) Mechanism 1. scRNA A detects mRNA detection target X (containing subsequence ‘a-b-c-d’) to form catalyst X·A, which mediates production of DsiRNA Dicer substrate B·C targeting mRNA silencing target Y (containing independent subsequence ‘v-w-x-y-z’). scRNAs A, B, and C coexist metastably in the absence of X. Successive toehold-mediated 3-way branch migrations enable assembly of X with A (step 1), X·A with B (step 2), X·A·B with C (step 3a), and disassembly of DsiRNA Dicer substrate B·C from catalyst X·A (step 3b). Domain lengths: |a| = 10, |b| = 10, |c| = 5, |d| = 2, |e| = 2, |v| = 2, |w| = 5, |x| = 2, |y| = 6, |z| = 5. Chemical modifications (2′OMe-RNA): A and parts of B and C (dashed backbone). (b) Conditional catalytic Dicer substrate formation. OFF state: minimal production of Dicer substrate B·C in the absence of detection target X, the presence of mRNA silencing target Y, or the presence of mRNA off-target Z. ON state: strong production of B·C in the presence of substoichiometric or stoichiometric short RNA detection target Xs (‘a-b-c-d’) or the presence of full-length mRNA detection target X. (c) Quantification of the Dicer substrate band (B·C) in panel (b). (d) Conditional Dicer processing. OFF state: minimal processing of the reactants (lane 2). ON state: efficient processing of Dicer substrate B·C (lane 4), yielding canonical 21- and 23-nt siRNAs (boxed bands). The non-siRNA remainder of the cleaved substrate is labeled ‘waste’. See Section S2 for additional computational and experimental studies of Mechanism 1.