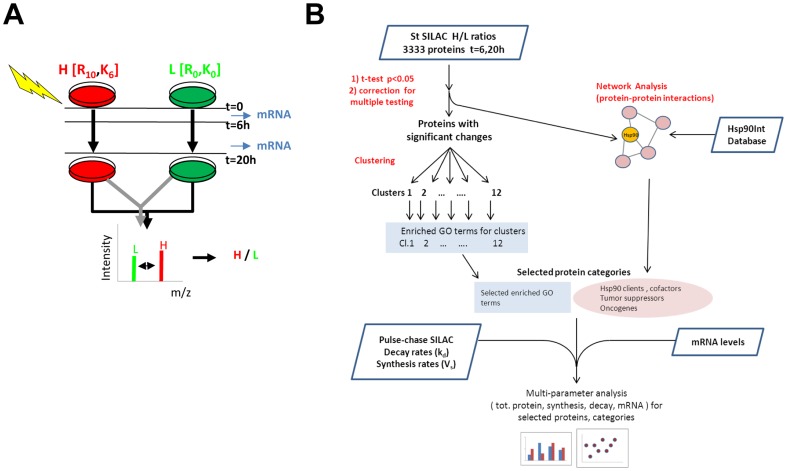
Figure 1. stSILAC experiments and workflow for data analysis.
A) Labelling and sample preparation scheme. Geldanamycin or DMSO were added at t = 0. Total protein extracts were collected at t = 6h and 20h, while total mRNA was taken at t = 5h and t = 19h. B) Data analysis and interpretation combined data on protein abundance changes (stSILAC) with protein-protein interactions (PPI) analysed as a network, synthesis and decay values for proteins in the two conditions and transcript levels. Enrichment of Gene Ontology annotation terms was used to extract functional information on protein categories with common behaviours.