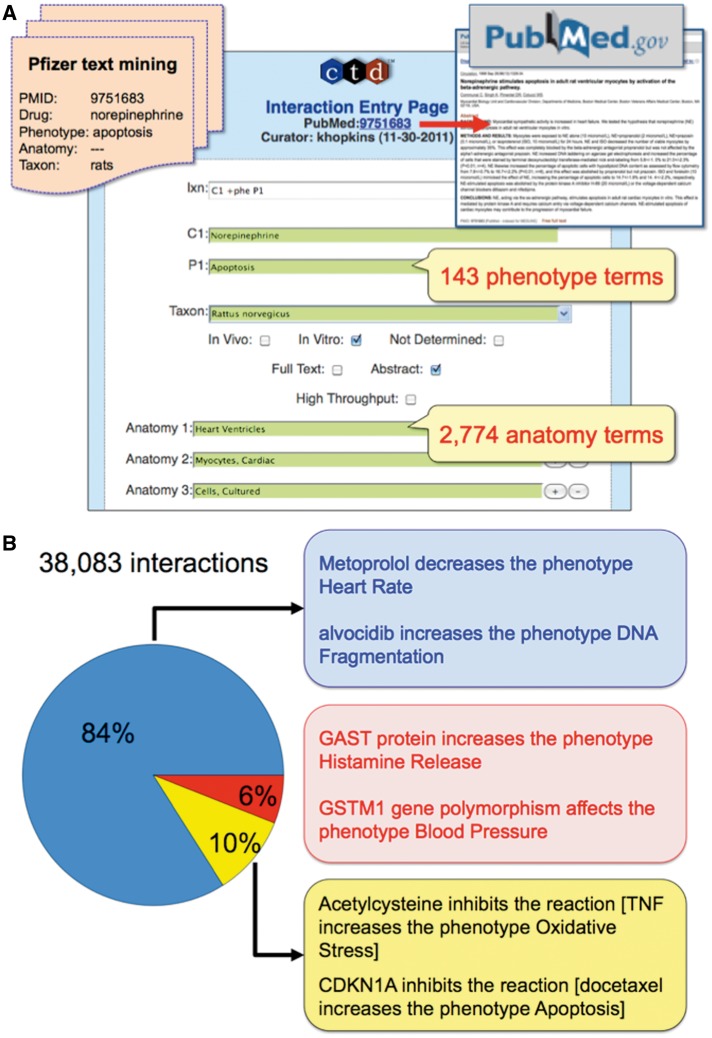
Figure 4.
CTD’s phenotype curation module. (A) Pfizer provided CTD with 10 366 articles text mined for a drug-of-interest, phenotype, anatomy and taxon (orange file, upper-left corner). Biocurators entered each article’s PMID into the CTD Curation Tool and retrieved the PubMed abstract for curatorial review (red arrow and box, upper-right corner). Biocurators curated from just the abstract whenever possible, but examined the full text if necessary to resolve any relevant issues mentioned in the abstract. Drug–phenotype interactions were generated using CTD’s structured notation, codes and controlled vocabularies in the Curation Tool (blue panel). In this prototype, 143 phenotype terms and 2774 anatomy terms were available. Here, the biocurator coded an interaction (Ixn field) describing how the drug norepinephrine (C1 field) resulted in increased apoptosis (P1 field) using an in vitro system from rats (Taxon field) of cultured ventricular myocytes (Anatomy 1–3 fields). The Curation Tool validates terms entered by the biocurator in real-time, and the green color of the text boxes indicates the terms are valid for curation. (B) Examples of CTD’s curated phenotype interactions. Of the total 38 083 interactions, 84% describe chemical–phenotype interactions (blue box), 6% gene–phenotype interactions (red box) and 10% complex chemical–gene–phenotype interactions (yellow box).