Abstract
Highly fragmented and morphologically indistinct fossil bone is common in archaeological and paleontological deposits but unfortunately it is of little use in compiling faunal assemblages. The development of a cost-effective methodology to taxonomically identify bulk bone is therefore a key challenge. Here, an ancient DNA methodology using high-throughput sequencing is developed to survey and analyse thousands of archaeological bones from southwest Australia. Fossils were collectively ground together depending on which of fifteen stratigraphical layers they were excavated from. By generating fifteen synthetic blends of bulk bone powder, each corresponding to a chronologically distinct layer, samples could be collectively analysed in an efficient manner. A diverse range of taxa, including endemic, extirpated and hitherto unrecorded taxa, dating back to c.46,000 years BP was characterized. The method is a novel, cost-effective use for unidentifiable bone fragments and a powerful molecular tool for surveying fossils that otherwise end up on the taxonomic “scrapheap”.
Fossil assemblages offer insights into past biodiversity, palaeoecology and human activities1,2,3. However, the accuracy of fossil identifications relies on the preservation of taxonomically significant morphological features, which are often lacking in highly fragmented remains. Over the past decade, analyses of ancient DNA (aDNA) have developed in sophistication and the breadth of contexts in which they are applied. Ancient DNA has been used to address questions of speciation, extinction and disease4,5,6,7 using a variety of substrates, including bone8, hair9 and eggshell10. However, to date, no study has attempted to use aDNA from taxonomically diverse fossils to map faunal assemblage data from a single site, largely due to the time and cost associated with generating aDNA sequences from each bone fragment.
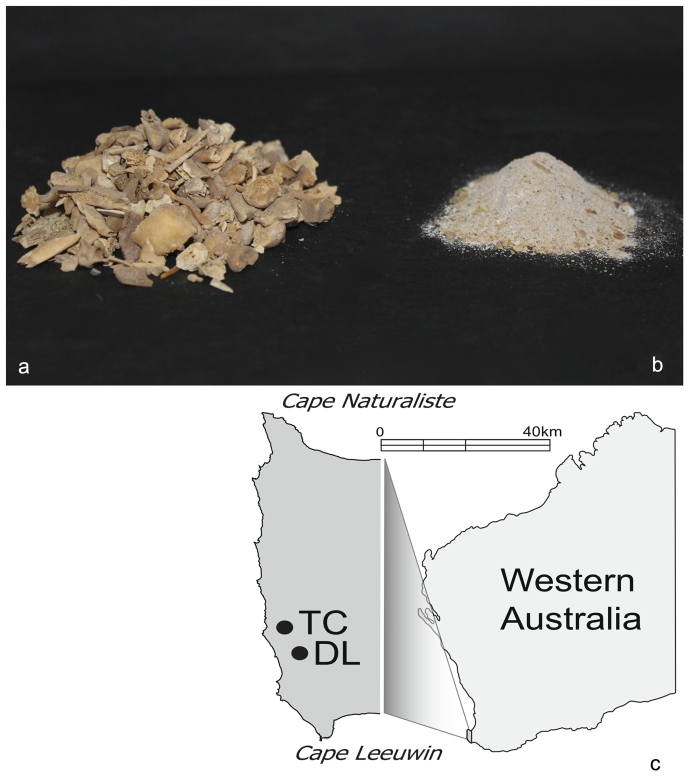
The destructive nature of sampling also means researchers and collection managers may be reluctant to analyse valuable specimens. At the same time, most archaeological and palaeontological excavations also collect large numbers of small, morphologically indistinct bone fragments (Figure 1a). Such material is of limited use in species identifications, although it may be important for some taphonomic analyses. Taxonomically, however, it is usually destined for the analytical “scrapheap”.
Figure 1. Bulk-bone fragments ground to form a bulk-bone powder at two archaeological sites.
Morphologically indistinct bulk-bone fragments (a) were ground to form single bulk-bone powder samples (b). Bulk-bone fragments were excavated from Devil's Lair (DL) and Tunnel Cave (TC), two archaeologically significant sites in southwest Western Australia (c). The map used in (c) was sourced from www.openclipart.org and was modified by J.H in Adobe Illustrator.
It is now possible, largely due to second generation high-throughput DNA sequencing (HTS) methodologies, to genetically profile complex, heterogeneous samples (Figure 1b) in parallel, both cheaply and quickly11,12. This DNA metabarcoding13 approach to genetically unravel complex substrates via HTS, as opposed to cloning, has transformed the analysis of substrates such as sediment14,15 and faecal material16,17. To explore large HTS-generated genomic datasets from environmental samples researchers use tools that are either: 1) taxonomy-dependent, which involves searching DNA reference databases for query and reference sequence matches18,19, or 2) taxonomy-independent, which involves taxonomy-independent measures of sequence diversity and clustering such as Operational Taxonomic Unit (OTU) analysis or UniFrac-based methods20,21,22.
This study seeks to employ HTS technology to sequence and identify aDNA obtained from thousands of morphologically unidentifiable archaeological bone fragments freshly excavated from deposits at Tunnel Cave (115° 02′ E, 34° 05′ S) and Devil's Lair (115° 04′ E, 30° 09′ S), two archaeologically and culturally significant sites in southwestern Australia (Figure 1c). Taken together, these sites, used to explore this methodological approach, span the last c.50,000 years23 and provide an unparalleled opportunity to study past Australian biodiversity and Aboriginal occupation23 located within an internationally recognised biodiversity “hotspot”24. A new method for the bulk sampling of fragmented bone material that would otherwise remain an untapped taxonomic resource is presented. By grinding multiple bones (Figure 1a) into an artificial “bulk-bone powder” (Figure 1b), thus producing a single bulk-bone powder sample, a large amount of highly informative genetic data can be quickly extracted. Such an approach should become commonplace in archaeological and palaeontological practice as it enables rapid assessment of DNA preservation and effectively maps zooarchaeological and palaeontological assemblages without destructive sampling of more valuable fossils.
Results
Overview of data generated
In a 2012 excavation, thousands of small bone fragments were collected by dry-sieving sediment from 15 well-dated stratigraphic units or layers at Devil's Lair and Tunnel Cave (Figure 1c). Around 50–150 bone fragments from within each layer were each drilled for 10–15s to form 15 bulk-bone powder samples representing the 15 layers (Figures 1a, 1b). DNA was extracted from each bulk-bone powder sample using established extraction methods (described in Methods) as if the bulk-bone sample were a single-source sample. The DNA extracts were screened for amplifiable mitochondrial DNA (mtDNA) using generic primers (tagged with HTS adaptors and unique barcodes) and subsequently sequenced using two HTS platforms: the GS-Junior (Roche) and the Ion Torrent PGM (Life Technologies).
Ancient DNA was successfully extracted from all bulk-bone powder samples, including a layer dated c.44,260–46,890 years BP (uncalibrated). The successful amplification and sequencing of DNA from all 15 layers was a rapid, cheap and effective way to assess DNA preservation at the sites (Figure 1c).
Amplicon DNA sequences (hereafter referred to as sequences) obtained from collective GS-Junior and Ion Torrent PGM sequencing runs were analysed for quality and possible chimeras. Except for ubiquitous human DNA sequences, control reactions throughout the process (described in Methods) were negative for contaminating DNA arising from laboratory processing.
Short regions within the mammalian mitochondrial 12S and 16S rRNA genes were amplified generating products of 100–104 bp and 90–96 bp respectively25. Amplification and sequencing of avian mtDNA was successful for some samples, producing either a 106–121 bp or 227–239 bp region of the avian mtDNA 12S gene25. Some cross-species reactivity was observed when using both 12S and 16S mammalian primer sets, resulting in the amplification and sequencing of avian and reptilian DNA. A targeted quantitative PCR (qPCR) and HTS approach to identify snake species was successful for a single sample.
Taxonomic identification
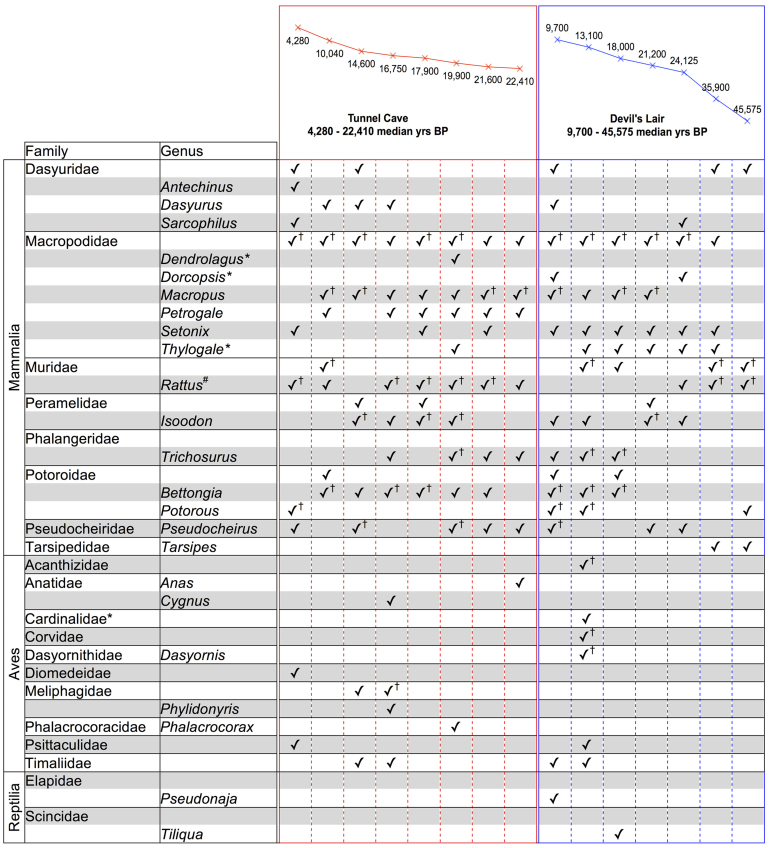
Mammalian 12S and 16S assays identified eight mammalian families representing 16 genera, using assignment filters chosen for this study (see Methods; Figure 2). The increase in sequencing depth afforded by the Ion Torrent PGM, as compared to the GS-Junior, did not increase the diversity of taxa identified. Mammalian taxa endemic to Australia were detected in multiple samples, in addition to taxa that have undergone significant range contraction and extirpation. The macropodid genus Thylogale (pademelon), provided the closest BLAST matches for many sequences across multiple samples, but to date no member of the genus has been recorded in this region. It was not possible to provide accurate taxonomic identifications for most of the Muridae sequences and for many Macropus sequences. While many sequences could be assigned with high confidence to a genus level, others could not be assigned beyond family or genus. A number of birds and reptiles were also identified and these have been collated at the family and genus level (Figure 2). While assignment to the species level is certainly possible in many instances a conservative approach is adopted here to showcase the approach.
Figure 2. Taxa identified in bulk-bone powder samples.
Mammals, birds and reptiles identified in each sample are listed. Samples are grouped according to site from youngest to oldest in years BP (uncalibrated), which is plotted on the same scale for both sites. The criteria used in taxonomic assignment are detailed in the Methods. Note that there is uncertainty surrounding taxonomy with regards to both Timaliidae and Cardinalidae (See Discussion). Key: †Detected using multiple primer sets; *Taxa not historically known to occur in the study region; #Sequences assigned to Rattus aligned closest to native Rattus Fuscipes (bush rat).
Genetic biodiversity analysis
A largely taxonomy-independent approach was adopted to examine fluctuations in observed genetic diversity over time at both sites. While the taxa identified using the GS-Junior and Ion Torrent PGM were mostly congruent, coverage dependent OTU inflation, arising from homopolymer sequencing error (see Methods; Discussion) was observed. A modified OTU analysis filter was designed to reduce the influence of HTS homopolymer sequencing error26,27, by employing distance-based metrics obtained from sequence alignments, giving rise to a new method referred to here as Distance-based Taxonomic Units (DTUs).
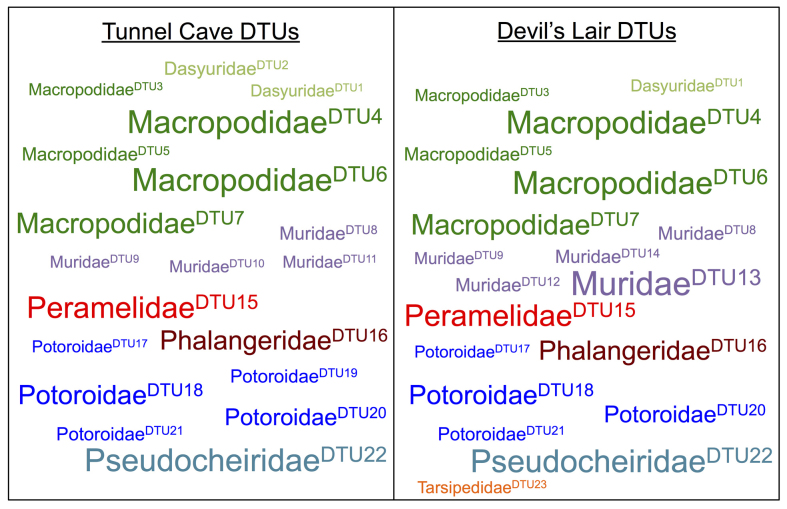
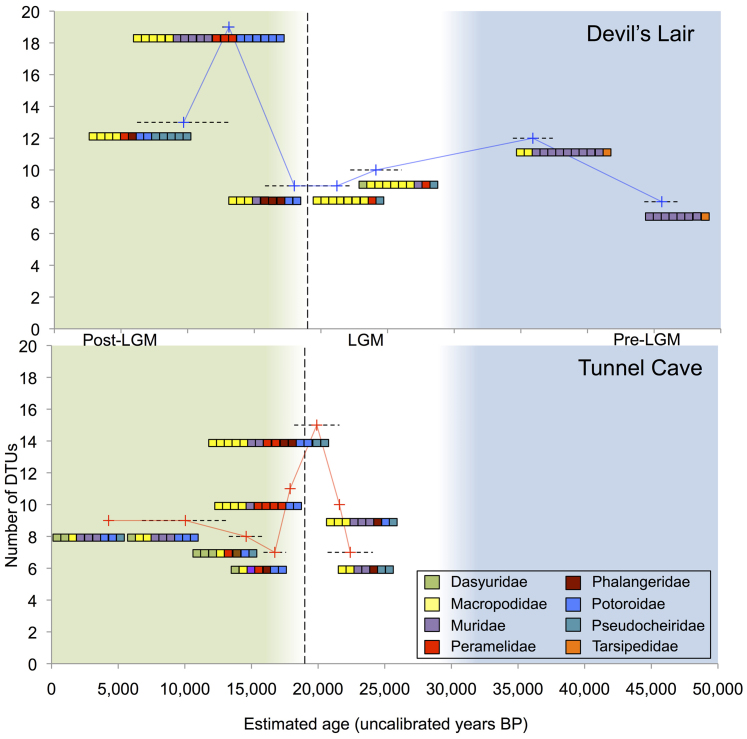
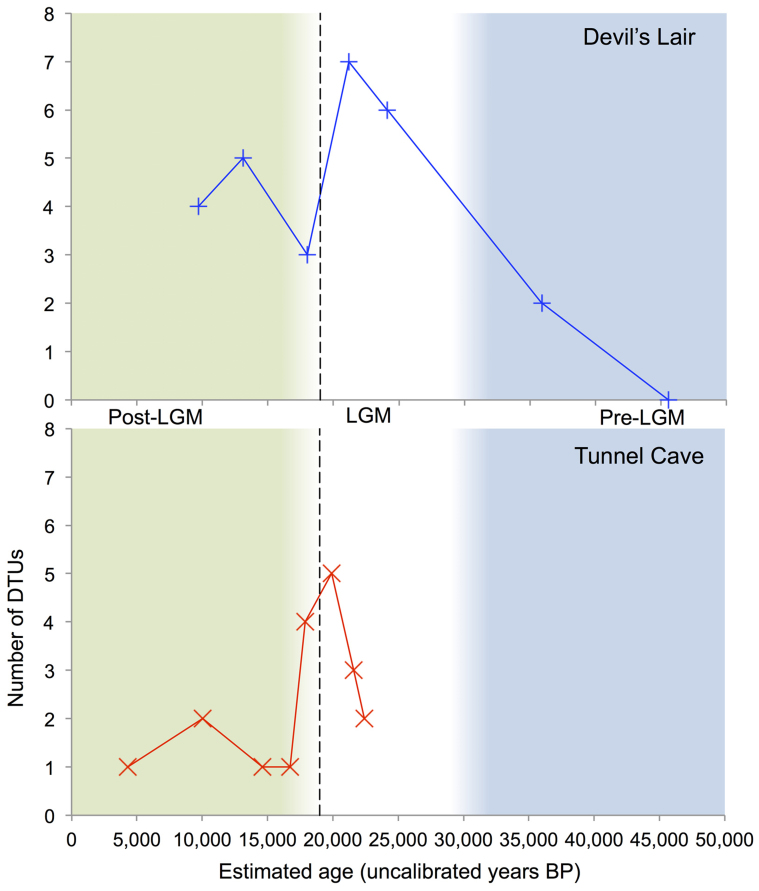
A total of 72 DTUs were identified across all 15 samples, 23 of which were shared across multiple samples, and in some instances both archeological sites (Figure 3). The number of DTUs fluctuates noticeably with time (Figure 4). The number of DTUs shows a notable decrease that roughly coincides with the last glacial maximum (LGM), whilst also showing an increase post-LGM. The composition of DTUs also varies over time. For instance, Potoroidae (potoroids) DTUs appear around the LGM and show an increase in numbers, whilst numbers of Macropodidae (macropodids) DTUs show a decline post-LGM.
Figure 3. DTUs shared across bulk-bone powder samples.
The DTUs shared between bulk-bone powder samples, and across both Tunnel Cave (left) and Devil's Lair (right), are shown. DTUs have been labeled with the closest BLAST family matches. Each DTU has been assigned a numeric identifier following the acronym ‘DTU’, shown in superscript. Font size is indicative of the total number of samples a DTU was detected in.
Figure 4. Change in DTU number and composition over time at Tunnel Cave and Devil's Lair.
The fluctuation in DTU number and the change in DTU composition across samples and at both sites are plotted against the backdrop of the major climatic shift around the end of the Last Glacial Maximum (LGM). Dashed vertical line - approximate end of the LGM; Blue background – Pre-LGM; White background – LGM; Green background – Post-LGM. Median ages are plotted for each sample; dashed horizontal line indicates minimum and maximum accepted date range for each layer.
With obvious variation in DTU composition, macropodid sequences were selected to examine DTU number flux at a finer scale to examine whether or not this reflected the overall trends in biodiversity change. Macropodids exhibit a declining trend in DTU diversity post-LGM (Figure 5) that marginally increases near the Holocene/Pleistocene transition 11,700 years ago.
Figure 5. Change in Macropodidae DTU number over time at Tunnel Cave and Devil's Lair.
The fluctuation in Macropodidae DTU number across samples and at both sites is illustrated. Dashed vertical line - approximate end of the LGM; Blue background – Pre-LGM; White background – LGM; Green background – Post-LGM. Median ages are plotted for each sample.
Discussion
This study presents a novel HTS method using aDNA characterised from bulk-bone powder samples. It represents a powerful new approach to analyse unidentifiable fragments excavated from fossil deposits. Ancient DNA extracted from bones within a layer dated between 44,260–46,890 years BP (uncalibrated), is the oldest aDNA recovered from Australia to date. These HTS results and the initial exploration of this technique show promise for larger scale bulk-bone analyses of fossil deposits. Rapidly analysing a bulk bone sample to determine if a site is conducive to DNA preservation will be valuable in excavations and test pits as DNA becomes increasingly incorporated into archaeological and palaeontological practices.
Even with the limited sampling, this first foray into bulk-bone analyses, has uncovered a significant amount of biological information that adds substantially to previous knowledge of the sites and surrounding biodiversity. Analysing these data in the context of DNA damage, sequencing error, incomplete reference databases and the necessary use of short DNA sequences raises numerous challenges that must be systematically addressed17,28,29,30. Nevertheless, when appropriate protocols and sequence filters are applied (see Methods) the method affords new insights into past biodiversity (Figure 2) and its temporal and spatial variation (Figures 3, 4 and 5).
Raw DNA sequences obtained from HTS platforms can be sorted and screened using a combination of filters that collectively exclude low-quality reads (Q-scores), sequences with errors in known flanking regions (adaptors, primers, and barcodes), artificial chimeric sequences and low abundance reads (see Methods). However, even sequences that pass these filters need to be interpreted with caution: the bird family Cardinalidae, which is not known to occur in Australia, is a case in point. The identification of birds also serves to illustrate the pitfalls associated with taxonomic revision. The taxonomy of the family Cardinalidae has been revised on a number of occasions, as has that of Timaliidae, which was also identified in some samples. Timaliidae has been regarded as a family consisting of Old World passerine birds, however the Australasian babblers (family: Pomatostomidae) were once within this family and the typical white-eyes (Zosterops) are disputably within this family also31. The families and genera identified (Figure 2) within each of the 15 samples require further investigation to identify taxa to the species level. Nevertheless, most of the genera identified at both sites from fossil morphology were again successfully detected in the bulk-bone23. The absence of some morphologically identified taxa from the genetically-determined faunal assemblage list is most likely due to sampling bias, as the present analysis derives from deposits representing less than one percent of the volume of the original excavations. Additionally, the possibility of primer binding bias contributing to the discontinuities between both aDNA and fossil assemblage datasets cannot be excluded. In silico analysis of variation in binding sites and the use of the multiple markers attempts to identify and minimize the impact of amplification bias. Finally, inherent differences between bones in terms of the preservation and quantum of mtDNA per unit biomass may also skew results between both methods of analysis causing artifactual over-representation of some taxa relative to others. However, taxa were also identified that were not detected in any previous morphology-based analyses, particularly small mammals, birds and reptiles, all of which require highly-specialised taxonomic skills to identify, are less likely to preserve diagnostic remains, and may be poorly represented in reference collections.
A high level of confidence surrounds the bulk of the taxonomic identifications; for instance, the majority of mammalian taxa identified are locally extant or known from the fossil record. The same generally holds true for avian and reptilian taxa identifications. The detection of sequences endemic to southwest Australia, such as a 100% match to Tarsipes rostratus (honey possum), further supports the bona fide nature of the sequences obtained. Moreover, the detection of extirpated taxa, such as Setonix (quokka) and Sarcophilus (Tasmanian devil), as far back as c.24,000 years BP (uncalibrated) illustrates the antiquity and authenticity of the sequences, as does the detection of species whose ranges have contracted and are no longer documented at the sites, e.g. Bettongia (bettongs). There appears to be little or no environmental contamination as evidenced by the absence of any sequences from highly abundant invasive taxa including Mus musculus (house mouse) or Rattus rattus (black rat). Whereas downward contamination may be an issue at some sites32, Devil's Lair contains several stratigraphical layers capped with calcite “flowstone”33 preventing the movement of fossils, and likely DNA5,23. Whilst it is acknowledged that contamination can be cryptic and sporadic34,35,36, the strict adherence to aDNA protocols37, the use of sequence quality filters and the plausibility of the data (see Methods), greatly reduces the likelihood that contamination contributed to the data presented here.
Although most taxonomic assignments from DNA sequences confirmed previous morphological identification23, some unexpected sequences resulted in distinct DTUs that were more difficult to assign. The issue is best exemplified by indeterminate Macropodidae sequences. It is unlikely that poor database coverage is the cause of this family-level assignment, as the Macropodidae database is nearly complete for both 16S and 12S rRNA mtDNA. In such cases sequencing error or DNA damage is also unlikely as the sequences are abundant and present across numerous samples at both sites, have passed all quality filters, form distinct DTUs and are unlikely to be nuclear copies (Figures 2, 3 and 4). It is possible therefore that these sequences may arise from extinct lineages of present-day macropodids or indeed from extinct taxa. In some cases sequences mapped closest to species of the New Guinea forest wallaby (Dorcopsis) and the east Australian restricted pademelon (Thylogale). The presence of such ‘indeterminate’ DNA sequences in bulk-bone samples is intriguing. For example, two extinct tree-kangaroo species (genus Bohra38,39), have been described in caves along the Nullarbor Plain, yet tree-kangaroos of the genus Dendrolagus are only currently present in northeastern Queensland and New Guinea and were previously not thought to have occurred so far south38. It is a tantalizing prospect that ‘indeterminate’ DNA sequences could represent previously unknown species from southwest Western Australia, but it is also a problematic finding, as there is no easy way to uncover the fossils that contributed the DNA. It is likely that bulk-sampling methods such as this will generate genetically plausible taxa that lack morphological identifications. Arguably a similar result has already occurred with the single Denisovan finger bone from “X-woman” used to postulate a new lineage of archaic humans in Siberia40,41.
When dealing with past biodiversity and aDNA sequences from fossil assemblages, analyses that are largely independent of taxonomy will likely be crucial to mapping temporal and/or spatial variation in genetic signatures. Such an approach facilitates the use of sequences that would otherwise be labeled “indeterminate”, which will be commonly encountered when employing the bulk-bone HTS methodologies advocated here. While it is not possible to comprehensively analyse changes in biodiversity over time presented here from only a handful of samples such an analysis serves to illustrate how bulk-bone data could be approached. The data presented in Figures 3,4,5 should therefore be viewed tentatively, as further extensive replication and investigation is required to confirm any significant patterning over time.
Owing to the difficulties of definitively assigning sequences to a defined taxonomy, a modified OTU analysis (referred to as DTU), has been introduced to examine biodiversity change over time. It was clear from the initial analysis that OTU numbers were artificially inflated primarily by homopolymer error. When dealing with short sequences homopolymer errors can create a distinct OTU whereby the only difference between it and its closest OTU match is a base within a homopolymer stretch. It was observed that homopolymer-derived OTUs were more common in those samples with greater depth of sequencing coverage. To overcome this issue, an OTU alignment and Kimura 2-parameter distance matrix was adopted whereby errors in homopolymer stretches appear as gaps and homopolymer-derived OTUs collapse into a single DTU (See Methods). Whilst at these particular sites, it is a challenge to disentangle the roles of climate, DNA decay and past anthropogenic influences; shifts in DTU composition appear at the LGM and at the Holocene-Pleistocene transition (Figures 4 and 5). Furthermore, specific Macropodidae DTU analysis showed a reduction in DTU diversity and abundance over time, with a drop in diversity around the LGM (Figure 5). With these tentative patterns of biodiversity being derived from only 15 DNA extractions it is easy to conceptualize how, with adequate sampling and appropriate genetic markers, a bulk-bone sampling method will facilitate detailed mapping of faunal changes over time. Moreover, the method is cheaper than single bone approaches42,43 while augmenting traditional morphological analysis.
The bulk-bone aDNA metabarcoding method used in this study presents a new, cost effective approach to identifying bulk quantities of morphologically indistinct bone fragments that otherwise end up in the taxonomic scrapheap. From modest amounts of sieved material across multiple layers at two study sites it was possible to detect equivalent diversity as described in previous morphological analyses23. While some taxa previously identified were not detected (most noticeably Macropus species), the converse was also true. This method is by no means an attempt to supplant traditional morphological approaches to taxonomic identification and analysis. Rather, it complements these approaches and by means of DTU analysis indicates changes in genetic diversity through time. Besides improving the identification of fossil assemblages the method allows researchers to rapidly assess the DNA preservation potential of freshly excavated material, which will vary from site to site. The approach will be equally applicable to archaeological and palaeontological sites, providing snapshots of past faunal diversity and human subsistence in both taxonomic dependent and independent ways. As such, it is anticipated that a bulk-bone approach will become a valuable part of the archaeological and palaeontological toolkit.
Methods
Sample collection and processing
Thousands of indistinct bone fragments were collected from both Tunnel Cave and Devil's Lair during excavations in February 2012. Approximately 150 L (0.15 m3) of sediment was analysed at both sites. Sediment was dry-sieved on site, using 2 mm and 5 mm sieves, and bagged according to well-defined and dated stratigraphical layers23. Each bagged sample was screened for bone fragments off-site, which were kept in groupings according to the layers in which they were found. Fifteen bulk-bone samples representing fifteen layers were processed: eight from Tunnel Cave, covering a period from 4,160–24,110 years BP (uncalibrated)23, and seven from Devil's Lair, covering a period from 6,200–46,890 years BP (uncalibrated)23. Small sections of the bones within each layer (typically 50–150 bones) were drilled (Dremel 114 drill bits) for a few seconds each and approximately equal amounts of drilled material from each bone fragment within a single layer was combined to form a “bulk-bone powder”. Owing to inherent differences in the amount of DNA per unit of biomass between species and differential DNA preservation between individual bones, over-representation of certain bone material in terms of DNA amplicon sequences is unavoidable.
DNA extraction and screening
All laboratory work was conducted in keeping with standard aDNA protocols28. Approximately 1 g of bulk-bone powder from each sample, including a blank extraction control, was digested overnight on a lab rotator at 55°C in 5 mL of digestion buffer containing: 2.5 mL EDTA (0.5 M), 0.1 mL Tris-HCL (1 M), 5 mg Proteinase K powder, 50 μL DTT (1 M), 50 μL SDS and made up to a final volume of 5 mL using EDTA. DNA digests were centrifuged at 6,000 rpm for 2 mins and the supernatant was concentrated to 50 μL using AMICON 30,000 MWCO columns (Millipore) as per the manufacturer's instructions. Each concentrate was transferred to a clean 2 mL eppendorf tube and PBi buffer (Qiagen) totalling 250 μL (i.e. 5× the volume of concentrate) was added. Each 300 μL PBi/concentrate mix was subsequently transferred to Qiagen silica spin columns and centrifuged at 13,000 rpm. Columns were washed with 700 μL of AW1 followed by AW2. A final dry spin at 13,000 rpm for 1 min followed. DNA was eluted from the columns in 60 μL EB with a 1 min incubation at room temperature prior to centrifugation at 13,000 rpm for 1 min.
Extracts were screened for amplifiable mtDNA using multiple primer sets via qPCR at three concentrations - undiluted, 1/10 and 1/50. Extracts were screened for mammalian mtDNA using 12SA/O and 16Smam primer sets, designed to amplify a small region within mammalian 12S and 16S mitochondrial genes respectively25,44. Extracts were also tested for avian mtDNA using 12SA/E and 12SA/H primer sets, designed to amplify a short and slightly longer overlapping region of the avian mitochondrial 12S gene respectively44. Finally, extracts were tested for snake mtDNA using the following primers: 12s_tRNA_F1_S AAAGTATAGCACTGAAAATGCTAA and 12s_R1_Snake GTTAGCCTGATACCGGCTCCG, designed to amplify a short region within the mitochondrial 12S gene. Each qPCR reaction was made up to a total volume of 25 μL, containing 1× PCR Gold Buffer (Applied Biosystems), 2.5 mM MgCl2 (Applied Biosystems), 0.4 mg/mL BSA (Fisher Biotech, Aus), 0.25 mM of each dNTP (Astral Scientific, Aus), 0.4 μM forward primer, 0.4 μM reverse primer, 0.25 μL AmpliTaq Gold (Applied Biosystems), 0.6 μL SYBR Green (1:2,000, Life Sciences gel stain solution) and 2 μL DNA extract. Quantitative PCR cycling conditions for the 12SA/O and snake 12S qPCR assays were as follows: initial heat denaturation at 95°C for 5 mins, followed by 50 cycles of 95°C for 30 s; 55°C for 30 s (annealing step); 72°C for 45 s followed by a 1°C melt curve and final extension at 72°C for 10 mins. Cycling conditions for 16Smam, 12SA/E and 12SA/H assays were the same as for the 12SA/O assay, except the annealing temperature, which was 57°C in each case. For each qPCR assay, DNA extraction, negative PCR reagent and positive DNA template controls were included.
DNA sequencing
DNA extracts that successfully yielded DNA of sufficient quality, free of inhibition, as determined by initial qPCR screening45, were prepared for amplicon sequencing. DNA extracts successful for all primer sets were sequenced on Roche's GS-Junior. Additional, separate, amplicon sequences were generated for extracts using mammalian 12SA/O and 16Smam primer sets for sequencing on Life Technologies' Ion Torrent Personal Genome Machine (PGM).
For each primer set, DNA extracts were assigned a unique DNA tag11. Each sample was tagged at both the 5′ and 3′ end of the target sequence using separate tags at both ends, resulting in a unique forward and reverse tag combination for each sequence. Independent tagged qPCRs for all samples, across all primer sets, were carried out in 25 μL reactions with reaction components and cycling conditions as described in ‘Methods: DNA extraction and screening’. Tagged qPCR amplicons were generated in triplicate and combined, thus minimizing the effects of PCR stochasticity on low-template samples, purified using Agencourt AMPure XP PCR Purification Kit (Beckman Coulter Genomics, NSW, Aus), as per manufacturer's instructions and eluted in 40 μL H2O. Purified amplicons were pooled to form separate sequencing libraries according to primer set used and sequencing platform. GS-Junior libraries were quantified using qPCR to determine an appropriate volume of library for sequencing (described in Murray et al. 2011). Each 25 μL reaction contained 12.5 μL ABI Power SYBR master mix (Applied Biosystems), 0.4 μM A-adapter primer, 0.4 μM B-adapter primer, 8.5 μL H2O and 2 μL pooled library, with the following cycling conditions: 95°C for 5 mins; 40 cycles of 95°C for 15 s, 56°C for 1 min followed by a 1°C melt curve. The appropriate library volume for use on the Ion Torrent PGM was determined using a Bioanalyser 2100 (Agilent). For each tagged qPCR assay, negative qPCR controls were included and if found to contain amplifiable DNA these qPCR amplicons were incorporated into the appropriate pooled sequencing library. All sequencing was performed as per manufacturer's instructions, with the use of 200 bp reagents and a 314 chip on the PGM.
Sequence identification
Amplicon sequence reads (hereafter referred to as sequences) were sorted into sample batches based on unique DNA tags. Identification tags and primers were trimmed allowing for no mismatch in length or base composition using Geneious v6.0.5 (created by Biomatters, available from http://www.geneious.com/). Batched and trimmed sequences from both GS-Junior and Ion Torrent PGM sequencing runs were combined according to sample and primer used. Each combined file was dereplicated, thus grouping sequences of exact identity and length, using USEARCH46. Dereplicated sequence files were searched for artificial chimeric sequences using the UCHIME de novo method47 in USEARCH and were removed, in addition to sequences occurring only once (i.e. singletons). The remaining sequences in each sample were subsequently clustered at an identity threshold of 97% using USEARCH with the most abundant sequence within each cluster selected as the representative sequence. To reduce noise associated with sequencing error, low abundant clusters, classed as those that occur at less than 1% of the total number of unique sequences when clustered at 100% sequence identity, were removed from the dataset. While the selection of a 1% cut-off is somewhat arbitrary, it should negate the possibility of clusters remaining that are the result of sequencing error. Additionally, the decision to class clusters as being in low abundance with respect to the total number of unique sequences (as opposed to total number of sequences or total number of sequences within the most abundant cluster) was made to minimize the effects of preferential DNA preservation and/or amplification. For each sample, every sequence assigned to the remaining clusters were queried against the NCBI GenBank nucleotide database using BLASTn48 in YABI49, enabling taxonomic identification. Sequences were searched without a low complexity filter, with a gap penalties existence of five and extension of two, expected alignment value less than 1e-10 and a word count of seven. The BLASTn results obtained were imported into MEtaGenome Analyzer v4 (MEGAN), where they were mapped and visualised against the NCBI taxonomic framework (min. bit score = 35.0, top percentage = 5%, min. support = 1)50. Sequences that were obviously the result of contamination (primarily human and cow) were eliminated from all subsequent downstream analysis steps.
Sequences that were truncated when queried against the NCBI GenBank nucleotide database were discarded from taxonomic analysis. Sequences with percentage similarity to a reference below 90% were discarded. Where sequence similarities were between 90–95% these were assigned to a family level, while those between 95–100% were assigned to a genus. Owing to the difficulties in assigning taxa beyond the genus level for some families, in addition to issues associated with characterizing past biodiversity that has been lost, species identifications were avoided in this particular study. Sequences that provided high percentage similarity to query references at a species level may or may not be bona fide, however with current insufficient data it is prudent to categorise these sequences cautiously. Where multiple taxa had equal percentage similarity scores to a query sequence, such sequences were moved higher up the taxonomic rankings.
While the validity of filters and hard percentage cut-offs are always debatable, those chosen in the analysis of this dataset seemed to afford the best balance when accounting for low template amounts and post-mortem damage on short aDNA fragments.
Genetic biodiversity analysis
Cognisant of the difficulties associated with assigning sequences to lower taxonomic levels, a modified form of OTU analysis was applied to the 16Smam sequences obtained in this study. This allowed changes in observed genetic diversity over time at both sites to be investigated independently of the above taxonomic classifications. Sequences within each sample were clustered at 97% identity, filtered and representative sequences were selected as detailed in Methods: Sequence Identification. Representative sequences within each sample were aligned in Geneious using MAFFT's G-INS-I algorithm and default parameters51. MAFFT alignments were imported into MEGA552 where a distance matrix between OTUs within a sample was calculated using a Kimura 2-parameter model53, with all positions containing gaps and missing data ignored. OTUs less than 3% divergent from each other were collapsed into a single DTU. This serves the purpose of reducing the influence of HTS homopolymer sequencing error26,27 by collapsing multiple homopolymer-derived OTUs into a single DTU, as errors in homopolymer stretches appear as gaps and are not included in the calculation of the distance matrix. Whilst this is first and foremost a largely taxonomic-independent analysis it is still nonetheless useful to identify coarsely to which family each DTU belongs, as this gives an idea of the diversity of DTUs within specific families. As such, all DTUs were searched against the NCBI GenBank nucleotide database using BLASTn48 to identify the family to which each DTU could be easily assigned. For the faunal specific Macropodidae DTU analysis the same method as above was followed except that only sequences assigned to Macropodidae were selected.
Author Contributions
D.C.M., M.B. and J.H. designed the experiments. D.C.M., J.H., N.W., D.H. and J.D. excavated and prepared samples. D.C.M., J.H., M.I.B., D.H. and R.A. contributed to HTS data generation and bioinformatics. J.D. provided stratigraphic interpretations and G.P. and J.D. provided fossil and taxon interpretations. D.C.M. and M.B. wrote the paper with assistance from all co-authors.
Acknowledgments
We thank the Wardandi people, and the Webb family in particular, as traditional owners and custodians of the region, for supporting our excavations and analyses. We thank student volunteers for their assistance in the field, Nina Kresoje (UWA) and Vanessa Atkinson (Pathwest Laboratory Medicine WA) and Frances Brigg for sequencing assistance. We also thank the WA Museum and the Curator of Anthropology Moya Smith for access to museum resources, and iVEC for computational support. Australian Research Council grants DP120103725 (to M.B., J.D. and J.H.) and FT0991741 (to M.B.) funded the research.
References
- Archibald S. B., Greenwood D. R. & Mathewes R. W. Seasonality, montane beta diversity, and Eocene insects: Testing Janzen's dispersal hypothesis in an equable world. Palaeogeogr. Palaeoclimatol. Palaeoecol. 371, 1–8 (2013). [Google Scholar]
- Colonesea A. C. et al. Holocene snail shell isotopic record of millennial-scale hydrological conditions in western Mediterranean: Data from Bauma del Serrat del Pont (NE Iberian Peninsula). Quat. Int. 303, 43–53 (2013). [Google Scholar]
- Dortch J. & Wright R. Identifying palaeo-environments and changes in Aboriginal subsistence from dual-patterned faunal assemblages, south-western Australia. J. Archaeol. Sci. 37, 1053–1064 (2010). [Google Scholar]
- Rohland N. et al. Genomic DNA Sequences from Mastodon and Woolly Mammoth Reveal Deep Speciation of Forest and Savanna Elephants. PLoS Biol. 8, e1000564 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haile J. et al. Ancient DNA reveals late survival of mammoth and horse in interior Alaska. Proc. Natl. Acad. Sci. U. S. A. 106, 22352–22357 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worobey M. et al. Direct evidence of extensive diversity of HIV-1 in Kinshasa by 1960. Nature. 455, 661–664 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raoult D. et al. Molecular identification by “suicide PCR” of Yersinia pestis as the agent of Medieval Black Death. Proc. Natl. Acad. Sci. USA. 97, 12800–12803 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. I. et al. Neanderthal DNA. Not just old but old and cold? Nature. 410, 771–772 (2001). [DOI] [PubMed] [Google Scholar]
- Bonnichsen R. et al. Methods of the study of ancient hair: Radiocarbon dates and gene sequences from individual hairs. J. Archaeol. Sci. 28, 775–785 (2001). [Google Scholar]
- Oskam C. L. et al. Fossil avian eggshell preserves ancient DNA. Proc. R. Soc. Biol. Sci. Ser. B. 277, 1991–2000 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binladen J. et al. The use of coded PCR primers enables high-throughput sequencing of multiple homolog amplification products by 454 parallel sequencing. PLoS ONE. 2, e197 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shokralla S., Spall J. L., Gibson J. F. & Hajibabaei M. Next-generation sequencing technologies for environmental DNA research. Mol. Ecol. 21, 1794–1805 (2012). [DOI] [PubMed] [Google Scholar]
- Taberlet P., Coissac E., Pompanon F., Brochmann C. & Willerslev E. Towards next-generation biodiversity assessment using DNA metabarcoding. Mol. Ecol. 21, 2045–2050 (2012). [DOI] [PubMed] [Google Scholar]
- Jørgensen T. et al. A comparative study of ancient sedimentary DNA, pollen and macrofossils from permafrost sediments of northern Siberia reveals long-term vegetational stability. Mol. Ecol. 21, 1989–2003 (2012). [DOI] [PubMed] [Google Scholar]
- Jørgensen T. et al. Islands in the ice: detecting past vegetation on Greenlandic nunataks using historical records and sedimentary ancient DNA Meta-barcoding. Mol. Ecol. 21, 1980–1988 (2011). [DOI] [PubMed] [Google Scholar]
- Deagle B., Chiaradia A., McInnes J. & Jarman S. Pyrosequencing faecal DNA to determine diet of little penguins: is what goes in what comes out? Conserv. Genet. 11, 2039–2048 (2010). [Google Scholar]
- Murray D. C. et al. High-throughput sequencing of ancient plant and mammal DNA preserved in herbivore middens. Quat. Sci. Rev. 58, 135–145 (2012). [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W. & Lipman D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- Little D. P. DNA barcode sequence identification incorporating taxonomic hierarchy and within taxon variability. PLoS ONE. 6, e20552 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 7, 335–336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss P. D. & Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71, 1501–1506 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M., Lozupone C. & Knight R. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. The ISME Journal. 4, 17–27 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dortch J. Palaeo-environmental Change and the Persistence of Human Occupation in South-western Australian Forests. (Archaeopress, Oxford, 2004). [Google Scholar]
- Myers N., Mittermeier R. A., Mittermeier C. G., de Fonseca G. A. B. & Kent J. Biodiversity hotspots for conservation priorities. Nature. 403, 853–858 (2000). [DOI] [PubMed] [Google Scholar]
- Taylor P. G. Reproducibility of ancient DNA sequences from extinct Pleistocene fauna. Mol. Biol. Evol. 13, 283–285 (1996). [DOI] [PubMed] [Google Scholar]
- Loman N. J. et al. Performance comparison of benchtop high-throughput sequencing platforms. Nat. Biotechnol. 30, 434–439 (2012). [DOI] [PubMed] [Google Scholar]
- Quail M. A. et al. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics. 13, 341 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A. & Poinar H. N. Ancient DNA: Do it right or not at all. Science. 289, 1139–1139 (2000). [DOI] [PubMed] [Google Scholar]
- Coissac E., Riaz T. & Puillandre N. Bioinformatic challenges for DNA metabarcoding of plants and animals. Mol. Ecol. 21, 1834–1847 (2012). [DOI] [PubMed] [Google Scholar]
- Taylor H. R. & Harris W. E. An emergent science on the brink of irrelevance: a review of the past 8 years of DNA barcoding. Mol. Ecol. Resour. 12, 377–388 (2012). [DOI] [PubMed] [Google Scholar]
- Jønsson K. A. & Fjeldså J. A phylogenetic supertree of oscine passerine birds (Aves: Passeri). Zoologica Scripta. 35, 149–186 (2006). [Google Scholar]
- Haile J. et al. Ancient DNA chronology within sediment deposits: Are paleobiological reconstructions possible and is DNA leaching a factor? Mol. Biol. Evol. 24, 982–989 (2007). [DOI] [PubMed] [Google Scholar]
- Turney C. & Bird M. I. Early human occupation at Devil's Lair, southwestern Australia 50,000 years ago. Quatern. Res. 55, 3–13 (2001). [Google Scholar]
- Champlot S. et al. An Efficient Multistrategy DNA Decontamination Procedure of PCR Reagents for Hypersensitive PCR Applications. PLoS ONE. 5, e13042 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlwein O. et al. DNA Extraction Columns Contaminated with Murine Sequences. PLoS ONE. 6, e23484 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuke P. W., Tettmar K. I., Tamuri A., Stoye J. P. & Tedder R. S. PCR Master Mixes Harbour Murine DNA Sequences. Caveat Emptor!. PLoS ONE. 6, e19953 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert M. T. P., Bandelt H.-J., Hofreiter M. & Barnes I. Assessing ancient DNA studies. Trends. Ecol. Evol. 20, 541–544 (2005). [DOI] [PubMed] [Google Scholar]
- Prideaux G. J. & Warburton N. M. A new Pleistocene tree-kangaroo (Diprotodontia: Macropodidae) from the Nullarbor Plain of south-central Australia. J. Vert. Paleontol. 28, 463–478 (2008). [Google Scholar]
- Prideaux G. J. & Warburton N. Bohra nullarbora sp. nov., a second tree-kangaroo (Marsupialia:Macropodidae) from the Pleistocene of the Nullarbor Plain, Western Australia. Rec. West. Aust. Mus. 25, 165–179 (2009). [Google Scholar]
- Krause J. et al. The complete mitochondrial DNA genome of an unknown hominin from southern Siberia. Nature. 464, 894–897 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M. et al. A high-coverage genome sequence from an archaic Denisovan individual. Science. 338, 222–226 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro B. et al. Rise and fall of the Beringian steppe bison. Science. 306, 1561–1565 (2004). [DOI] [PubMed] [Google Scholar]
- Lorenzen E. D. et al. Species-specific responses of Late Quaternary megafauna to climate and humans. Nature. 479, 359–364 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A. et al. Complete mitochondrial genome sequences of two extinct moas clarify ratite evolution. Nature. 409, 704–707 (2001). [DOI] [PubMed] [Google Scholar]
- Bunce M., Oskam C. & Allentoft M. in Ancient DNA: Methods and Protocols Methods in Molecular Biology (eds Shapiro, B. & Hofreiter, M.) Ch. 16, 121–132 (Humana Press, New York City, 2011). [Google Scholar]
- Edgar R. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 26, 2460–2461 (2010). [DOI] [PubMed] [Google Scholar]
- Edgar R. C., Haas B. J., Clemente J. C., Quince C. & Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 27, 2194–2200 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson D. A., Karsch-Mizrachi I., Lipman D. J., Ostell J. & Wheeler D. L. GenBank. Nucleic Acids Res. 34, D16–D20 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter A. A., Macgregor A. B., Szabo T. O., Wellington C. A. & Bellgard M. I. Yabi: An online research environment for grid, high performance and cloud computing. Source Code Biol. Med. 7, 1 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson D. H., Auch A. F., Qi J. & Schuster S. C. MEGAN analysis of metagenomic data. Genome Res. 17, 377–386 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Misawa K., Kuma K. & Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K. et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120 (1980). [DOI] [PubMed] [Google Scholar]