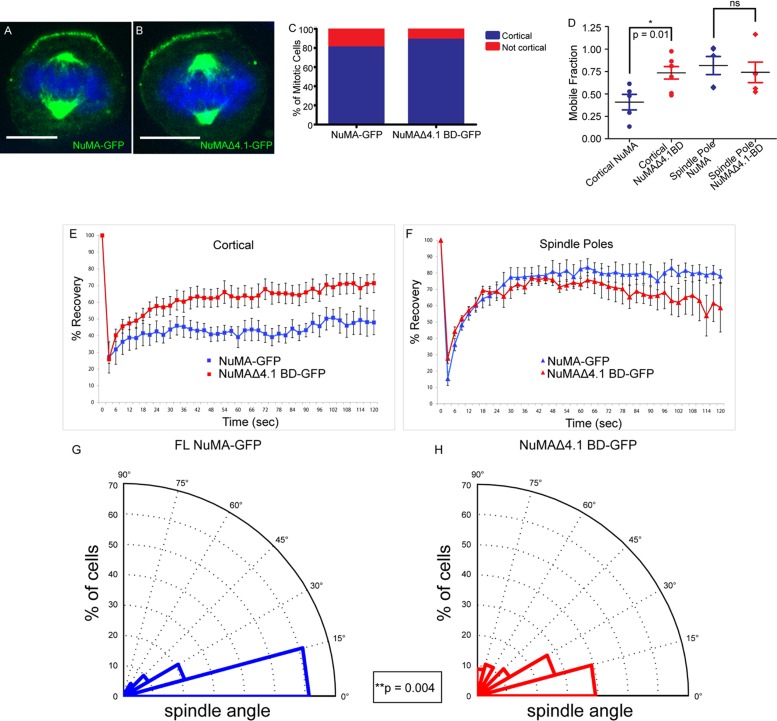
FIGURE 3:
4.1/NuMA interactions are required for cortical NuMA stability and mitotic spindle orientation. (A, B) Full-length NuMA-GFP and NuMAΔ4.1 BD-GFP constructs were transfected into wild-type cells, and immunofluorescence analysis was performed to determine localization. (C) Quantitation of cortical accumulation of GFP-tagged NuMA and NuMAΔ4.1 BD. n = 25 cells for each, p = 0.7. (D) Dot plot showing the distribution of mobile fractions of cortical and spindle pole NuMA-GFP for both WT and NuMAΔ4.1 BD. p (cortical) = 0.01; p (spindle poles) = 0.63. (E, F) FRAP analysis of full-length NuMA-GFP (FL) vs. a GFP-tagged NuMA construct lacking the 4.1-binding domain (Δ4.1 BD) transfected into wild-type cells. Recovery profiles of bleached GFP cortical signal (E) and spindle pole signal (F) from both constructs over 2 min. (G, H) Analysis of mitotic spindle alignment with the center of the cortical NuMA-GFP crescent in FL and NuMAΔ4.1 BD-GFP–transfected wild-type cells plated on 100 μM laminin. Radial histograms illustrate the distribution of spindle angles from both constructs. Histograms are grouped into 15° bins, with 0° representing a spindle aligned with the center of the NuMA crescent. n = 62 for NuMA-GFP, n = 56 for NuMAΔ4.1 BD-GFP; p = 0.004 using a Kolmogorov–Smirnov test. Scale bars, 10 μm.