FIGURE 6:
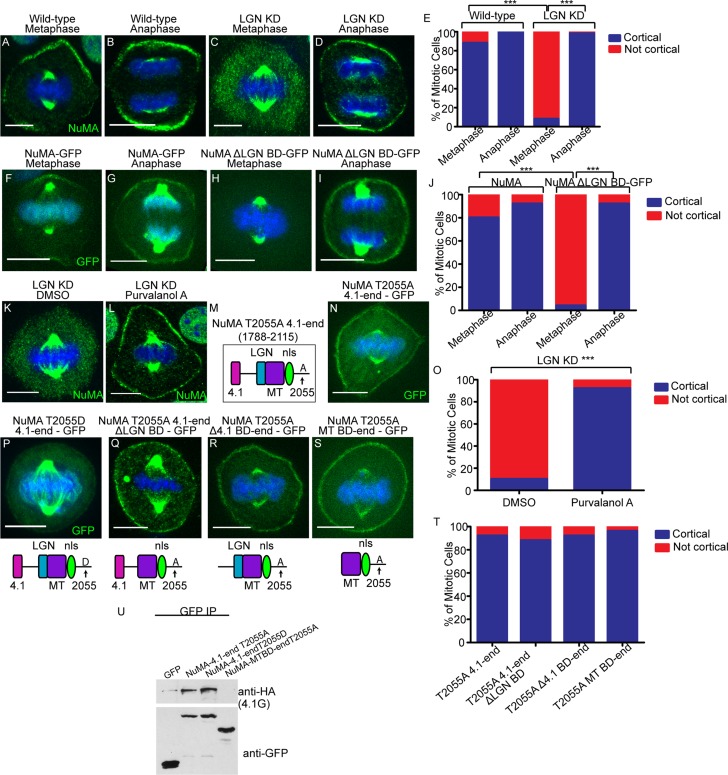
Anaphase localization of NuMA is independent of LGN and 4.1 binding. (A–D) Immunofluorescence analysis of endogenous NuMA localization in wild-type and LGN-knockdown keratinocytes in metaphase and anaphase, as indicated. (E) Quantitation of cells with cortical NuMA accumulation, as indicated. n = 50 cells, p < 0.0001 for NuMA localization in WT vs. LGN KD cells in metaphase. (F–I) Epifluorescence of NuMA-GFP and NuMAΔLGN–BD-GFP transfected keratinocytes in metaphase and anaphase, as indicated. (J) Quantitation of cells with cortical NuMA accumulation, as indicated. n = 25 cells, p < 0.0001 for NuMA-GFP vs. NuMAΔLGN BD-GFP cortical localization in metaphase. (K, L) Immunofluorescence analysis of endogenous NuMA localization in LGN-knockdown keratinocytes treated with DMSO or 100 μM purvalanol A for 3 min. (M) NuMA construct that harbors the T2055A mutation and spans from the carboxy-terminal 4.1-binding domain to the end of the protein. (N) A GFP-tagged version of the NuMA fragment diagrammed in M was expressed in wild-type cells, and immunofluorescence analysis was performed to determine localization. (O) Quantitation of cells from K and L with cortical NuMA accumulation, as indicated. n = 50 cells, p < 0.0001. (P–S) GFP epifluorescence of NuMA constructs as indicated. The domain structure of each NuMA fragment is diagrammed beneath its corresponding image. (T) Quantitation of the ability of NuMA deletion/mutant constructs from N and Q–S to target to the cell cortex in metaphase. n = 25 cells, all p > 0.05. (U) Immunoprecipitation of GFP-tagged NuMA constructs/mutants with 4.1G-HA. Top and bottom, amount of 4.1G-HA and GFP fusion proteins in the immunoprecipitates, respectively.