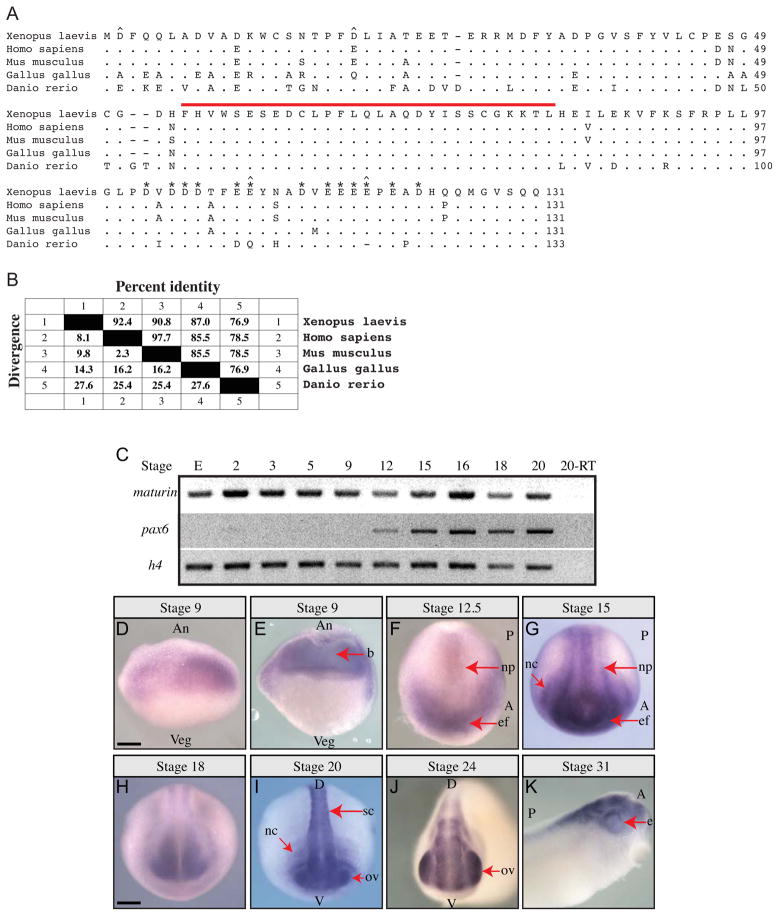
Fig. 1.
Maturin predicted protein sequence, conservation and expression during Xenopus laevis embryonic development. (A) Maturin protein sequence alignment. Predicted amino acid sequence of X. laevis Maturin aligned with putative orthologs from other vertebrates. Periods indicate amino acid identity, while gaps (dashes) were inserted to optimize the alignment. The 29-residue Maturin Motif is indicated by a red over-line. Aspartic (D) and glutamic (E) amino acids in the acidic domain are labeled with asterisks. The four positions at which nonconservative changes of highly acidic amino acids have taken place are indicated with a caret (^). (B) Amino acid identity among vertebrate Maturins. Percent identity shared with X. laevis Maturin is shown. Genbank accession numbers are: X. laevis BC045253, H. sapiens NM_152793 (C7orf41), M. musculus BC042507 (2410066E13Rik), G. gallus XM_003640720.1, D. rerio NM_001144806. (C) Detection of maturin and pax6 transcripts using RT-PCR. Gene specific primers were used to detect maturin and pax6 transcripts in total RNA isolated from eggs (E) and whole embryos of the indicated developmental stage. Histone h4 was used as a loading control. (D–K) Maturin expression in developing embryos. Whole mount in situ hybridization was used to detect Xenopus laevis maturin expression. Developmental stage is indicated above each panel. (E) Stage 9 embryo cut open with a razor following whole mount in situ hybridization. An, animal; Veg, vegetal; b, blastocoel; A, anterior; P, posterior; np, neural plate; ef, eye field; nc, neural crest; D, dorsal; V, ventral; sc, spinal cord; ov, optic vesicle; e, eye. Scale bars = 400μm.