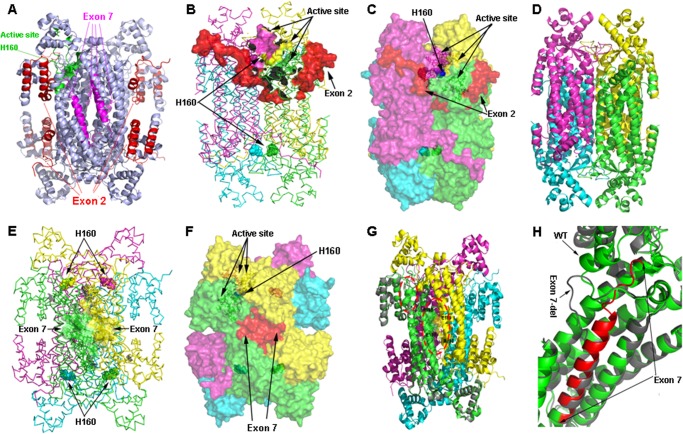
FIGURE 6.
Tetrameric structure of wild type and models of ASL exon 2- or exon 7-deleted transcript variant. A, tetrameric structure of WT ASL showing the positions of active site histidine 160 and exons 2 and 7. The structure is shown as a ribbon model in light blue; amino acids contributed by exon 2 are colored in red, whereas amino acids contributed by exon 7 are colored in magenta. The active site residues are colored in green with His-160 at the catalytic center shown a sphere model. B, location of exon 2 in relation to the active site. In the ASL homotetramer, active site residues are contributed by three different subunits. The ASL structure is shown as a ribbon model with different subunits colored in green, cyan, magenta, and yellow. The green and cyan subunits form a dimer and join with another dimer formed by magenta and yellow subunits to form the tetramer. Active site residues from one of the sites are shown as solid surfaces, whereas the catalytic center (His-160) is shown as spheres. Residues contributed by exon 2 are shown in red. C, role of the N terminus tail in stabilizing the active site. The ASL homotetramer is shown as a solid surface model with different subunits colored in green, cyan, magenta, and yellow. Residues contributed by exon 2 are shown in red. The N terminus tail surrounds the active site and provides the stability required for binding of substrate. D, conjectural prediction of exon 2-deleted ASL based on homology modeling. The core structure of the ASL homotetramer can still be formed in the absence of N terminus residues contributed by exon 2. The model of the ex2del tetrameric complex is similar to the WT ASL, and the central four-helix bundle comprising the tetramer remains intact, resulting in a stable structure. However, the loss of the N terminus required for substrate binding will result in an enzymatically inactive protein. E, the WT ASL tetramer structure showing the locations of residues contributed by exon 7. The homotetramer structure is shown as a ribbon model with different subunits colored in green, cyan, magenta, and yellow. The amino acids contributed by exon 7 are shown as surface models in two of the adjacent dimeric structures that form the tetramer. Chains shown in green and yellow are from two different dimers that join together to form the tetramer. The His-160 residue at the catalytic center is shown as spheres. F, a space-filled surface model of homotetrameric ASL structure showing the role of exon 7 in tetramer formation. Interactions between charged residues on adjacent dimeric ASL subunits stabilize the tetrameric structure. G, a structural model of exon 7 deleted variant of ASL transcript. One subunit of the exon 7-deleted variant is superimposed on the WT ASL homotetramer to show the structural differences. The protein is shown as a ribbon model with the exon 7-deleted variant colored in gray, whereas WT subunits in the tetramer are colored in green, cyan, magenta, and yellow. The residues involved in charge-based interaction located on two adjacent subunits in the WT protein are shown as sticks in the larger oval red dotted box. The secondary structure elements altered in the exon 7-deleted variant are marked in a smaller oval box. H, a close-up of the structural changes resulting from the exon 7 deletion. The exon 7-deleted variant (gray) and WT (green) are superimposed, and residues contributed by exon 7 in the WT protein are shown in red. Two of the central helices in the WT protein are altered to flexible loops in the exon 7-deleted variants, resulting in an unstable protein due to the requirement of core helices in domain 2 of the ASL monomers for overall stability. The resultant protein was found to be highly unstable and unlikely to participate in tetramer formation.